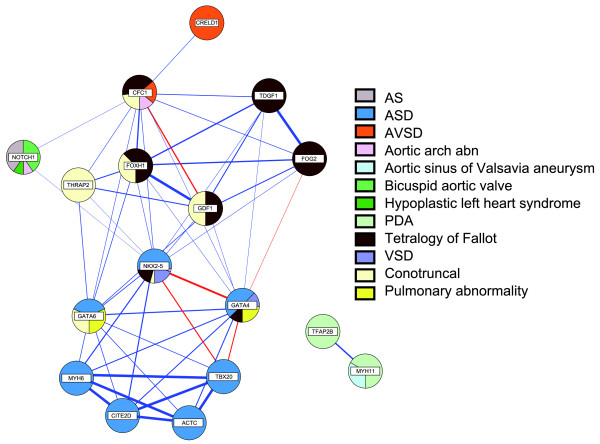
Figure 3.
Network of genes (nodes) sharing non-syndromic cardiac phenotypes when mutated (edges). The nodes are represented as pie charts displaying gene-linked CHD types as well as their frequency. Unconfirmed gene-phenotype relations based on single case reports were not included. The width of the edges (log of euclidian distance) depends on the number and the percentage of shared phenotypes. Known protein-protein interactions are represented by red edges. Proteins sharing multiple phenotypes when mutated tend to act in the same molecular pathways or even encode proteins that directly interact (for example, TBX20 physically interacts with NKX2.5 and GATA4 [33]. By further expanding the database, phenotype sharing will enable us to predict novel protein interaction partners. On the other hand, we hypothesize that further insights into the molecular basis of the developing heart will point towards novel candidate genes for a specific CHD type based upon the phenotypic spectrum of a known interaction partner. For example, since mutations in CFC1 are associated with laterality defects and conotruncal heart defects, other players in the NODAL signaling pathway (FOXH1, TDGF1 and GDF1) were considered to be likely candidates for these CHDs [34,35]. AS, aortic stenosis; ASD, atrials septal defect; AVSD, atrioventricular septal defect; PDA, patent ductus arteriosus; VSD, ventricular septal defect.