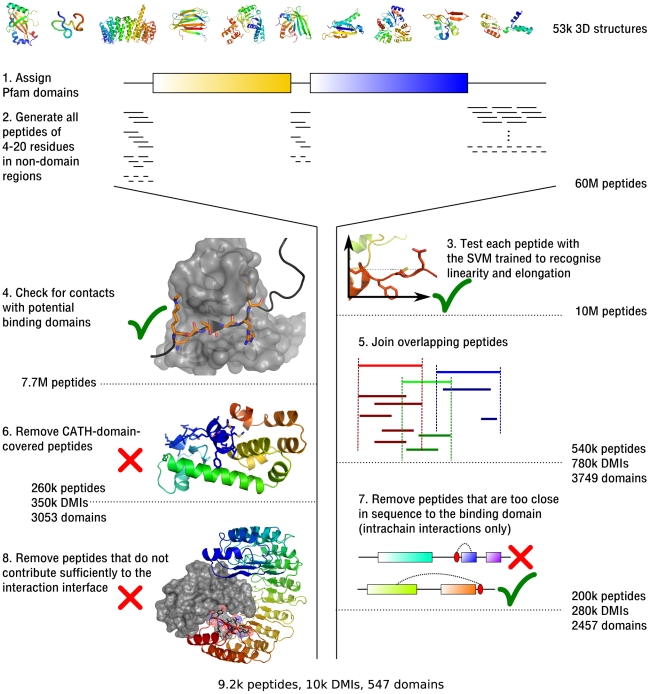
Figure 2. Overview of the generation and filtering of motif-like peptides.
(Steps 1 and 2) We generated all possible peptides of 4–20 residues from regions of 3D structures that did not match Pfam domains. (3, 4) For peptides accepted by the SVM trained on linearity and elongation (cf. Figure 1) we computed whether there were sufficient contacts with domains in the same structure, which may be in the same or in another protein chain. (5) Peptides that are completely covered by other (longer) peptides are removed, so that the largest accepted peptide represents shorter candidates binding to the same region. (6) Peptides in intrachain interactions that are part of CATH domains are often artefacts of differences between structure- and sequence-based domain assignment and are therefore excluded. (7) Peptides in intrachain interactions that are sequentially directly next to the binding domain are often artefacts and thus removed, though in general peptides close to domains are allowed, as long as they have a sufficient sequential distance from their binding domain. (8) Exclude candidate DMI in which the interface is smaller than 150 Å, or in which the interface between domain and peptide is less than 50% of the total interface between the proteins.