Fig. 3.
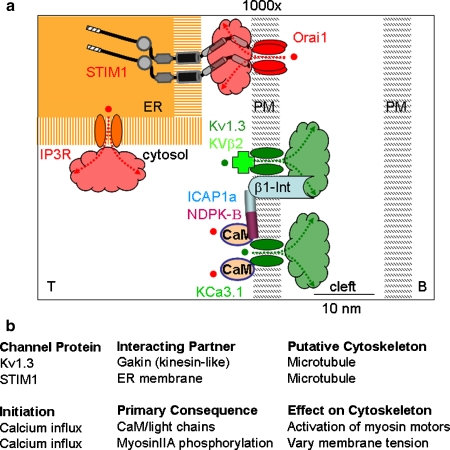
Calcium channels and the cytoskeleton at the immunological synapse. a Zoomed-in schematic diagram of region shown by box in the lower right panel of Fig. 2a, showing T-cell molecules concentrated at the synapse, separated by a narrow synaptic cleft (∼15–20 nm) from the B cell. Scales are approximate. A STIM1 dimer within the endoplasmic reticulum (ER) is shown activating a tetramer of Orai1 in the plasma membrane by direct contact. Protein domains of STIM1 include (from left to right): signal peptide, EF hand (without bound Ca2+), and SAM domains in the ER lumen; a transmembrane segment, and bipartite coiled-coil in cytosol. Depletion of the ER Ca2+ store by release of Ca2+ ions (red) through the IP3 receptor (IP 3 R) initiates signaling via STIM1 to the PM. Ca2+ first unbinds from the EF hand; STIM1 then forms oligomers that translocate to ER–PM junctions; Orai1 tetramers then open; Ca2+ ions enter the cell and begin to diffuse. Ca2+ opens KCa3.1/CaM; membrane depolarization opens Kv1.3. K+ ions (green), leaves the cytosol, and enters the cleft. Kv1.3 and KCa3.1 are linked through associated subunits to enzymes, to β1-integrin (β1-Int), and to cytoskeletal elements. Clouds indicate diffusion of Ca2+ and K+ ions and accumulation in nanodomains near Orai1 (red) and Kv1.3 and KCa3.1 channels (green). b Putative interplay between channels and the cytoskeleton. Channels themselves are likely assembled as a function of dynamic reorganization of cytoskeletal elements. Conversely, calcium transients and gradients almost certainly integrate at multiple levels to continue to stabilize or destabilize motility modes