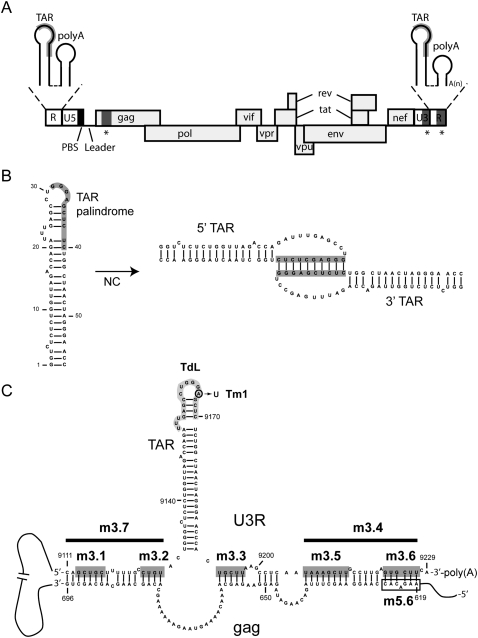
FIGURE 1.
(A) Schematic representation of the HIV-1 viral RNA genome. The positions of the open reading frames are indicated. The coding region is flanked at the 5′ and 3′ ends of the genome by untranslated leader regions (UTR). The 5′ UTR contains the repeat (R) region, the U5 region, and the leader sequence (L). The primer binding site (PBS) is marked by a black box and the TAR and poly(A) hairpins in the R region are shown as insets above. The 3′ UTR contains the U3 region and the R region. The sequences involved in the proposed TAR–TAR interaction are marked in gray, and the sequences involved in the proposed gag–U3R interaction are marked by asterisks. (B) Model for the TAR–TAR interaction (Andersen et al. 2004). The palindromic sequence that was found to induce dimerization, dependent on the nucleocapsid (NC) protein, is marked by a gray box. (C) Model for the gag–U3R interaction (Ooms et al. 2007). Sequences in the gag open reading frame (positions 619–696) were proposed to interact with sequences flanking the TAR hairpin in the U3R region. Mutations introduced in the U3R region are indicated. The sequences that were substituted in mutants m3.1, m3.2, m3.3, m3.5, and m3.6 are marked by dark gray boxes. The deletions introduced in mutants m3.4 and m3.7 are marked by a black line. Mutant Tm1 contains one substitution, A9166U, in the TAR hairpin. The sequence in the TAR hairpin marked in light gray was deleted and replaced by three A residues in mutant TdL. An open box marks the gag sequence substituted in mutant m5.6.