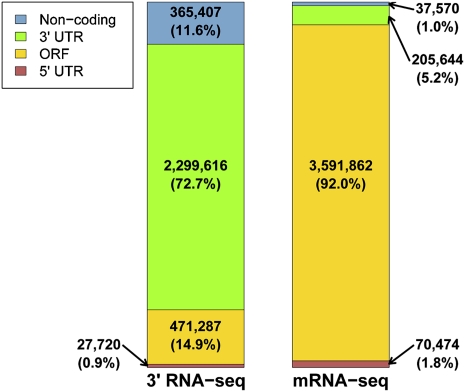
FIGURE 2.
The genomic distribution of mapped reads differs between 3′-end RNA-seq and standard mRNA-seq. Shown are the frequencies of uniquely mapped reads whose positions fell into the indicated genomic elements. Left, P1 3′-end RNA-seq library constructed as in Figure 1; right, mRNA-seq library constructed as in Nagalakshmi et al. (2008). For 3′-end RNA-seq, poly(A) (or poly(T)) positions were classified according to annotations on the same strand, while for mRNA-seq, start positions were classified without strand specificity. 5′ UTR definitions were taken from Nagalakshmi et al. (2008) and 3′ UTR lengths are defined in Materials and Methods. Noncoding, reads mapping to regions outside known ORFs, 5′ and 3′ UTRs, and known structural or regulatory RNAs.