Figure 1. Cyclin D1 regulation in normal versus cancer cells.
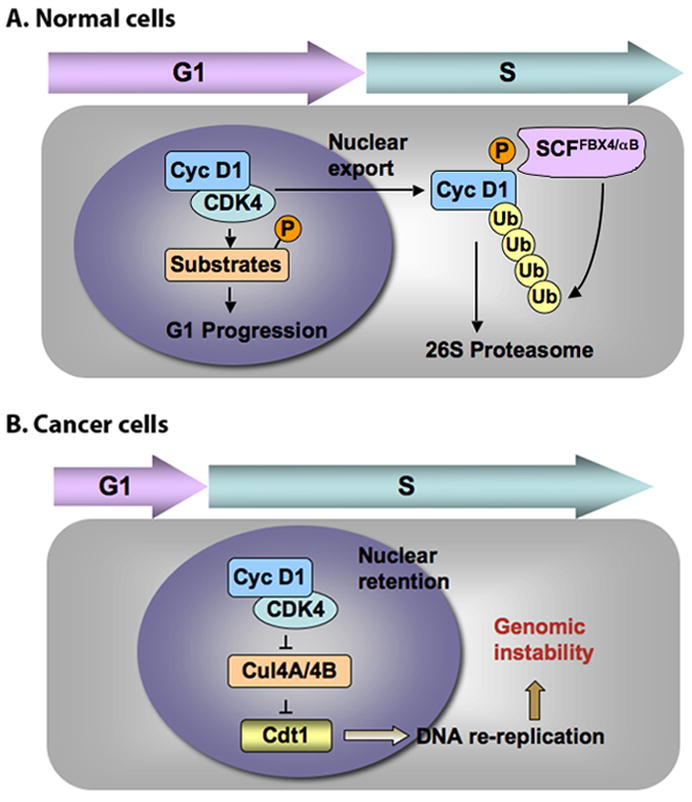
A: In normal cells, following G1 progression, cyclin D1 exits the nucleus via a distinct mechanism that involves GSK3β-mediated phosphorylation of Thr286 and CRM1-dependent nuclear export. Once in cytoplasm, phosphorylated cyclin D1 is recognized and ubiquitylated by SCFFbx4/αB-crystallin ligase, targeting the cyclin D1 to the 26S proteasome for degradation. B: In cancer cells, constitutive nuclear accumulation of active cyclin D1/CDK4 complexes can be achieved through one of several mechanisms: 1) mutations in cyclin D1 that prevent recognition by Crm1 (Benzeno et al., 2006; Moreno-Bueno et al., 2003); 2) mutation of Fbx4 which blocks ubiquitin-dependent degradation (Barbash et al., 2008); 3) alternative splicing of the cyclin D1 transcript which removes sequences that direct nuclear export and Fbx4-dependent destruction (Lu et al., 2003). The accumulation of active nuclear cyclin D1/CDK4 complex during S-phase stabilizes Cdt1 by transcriptionally inhibiting Cul4A and Cul4B, promoting DNA re-replication during S phase and consequently inducing genomic stability (Aggarwal et al., 2007).
