Abstract
A genetic basis for atopic dermatitis (AD) has long-been recognized. Historic documents allude to family history of disease as a risk factor. Prior to characterization of the human genome, heritability studies combined with family-based linkage studies supported the definition of AD as a complex trait, in that interactions between genes and environmental factors and the interplay between multiple genes contribute to disease manifestation. A summary of over 100 published reports on genetic association studies through mid- 2009 implicates 81 genes, 46 of which have demonstrated at least one positive association with AD. Of these, the gene encoding filaggrin (FLG) has been most consistently replicated. Most candidate gene studies to date have focused on adaptive and innate immune response genes, but there is increasing interest in skin barrier dysfunction genes. This review examines the methods that have been used to identify susceptibility genes for AD, and how the underlying pathology of this disease has been used to select candidate genes. Current challenges and the potential impact of new technologies are discussed.
Keywords: atopic dermatitis, genetics, IgE-mediated response, innate immunity, skin barrier dysfunction, genetic association, gene-environment interaction, ethnicity
INTRODUCTION
Over 2,500 years ago, Hippocrates described a condition of an undetermined cause characterized as “itching over [a patient’s] whole body”1. During the 18th and 19th centuries, what was believed to be this non-descript pruritic state was termed ‘eczema’, or “prurigo diathésique”2. The condition received increasing attention in the dermatologic literature in the late 19th and early 20th centuries, especially as a component of the allergic diathesis, as Coca noted that “every recognized category of allergic disease affects the skin”, suggesting that, “among the diseases of the skin the familial allergy (hay fever-asthma group) is represented by atopic eczema”3. The term was ultimately replaced with the descriptive term ‘atopic dermatitis’ by Coca, Sulzberger and Wise3.
A common theme from the most ancient descriptions of this syndrome to more recent observations is that allergy in general, and atopic dermatitis (AD) in particular, has a profound capacity to run in families. That is, AD, even more so than other atopic disorders, is highly ‘heritable’. For example, in the Munich Asthma and Allergy Study, it was demonstrated that the risk of a child developing AD if one or both parents have AD is higher (OR=3.4, 2.6–4.4) compared to the risk if one or both parents have asthma (OR=1.5, 1.0–2.2) or allergic rhinitis (OR=1.4, 1.1–1.8)4 supporting the notion that, although generic, ‘atopy’ genes may be responsible for manifestation of AD, there are likely to be phenotype-specific genes as well. Early observations of a strong family history of allergy, eczema and asthma5, 6 contributed not only to an understanding of at least one of the underlying ‘causes’ of AD – that is, an allergic response to “environmental inhalant factors” in addition to “hereditary factors”5 - but also promoted the next stage of heritability investigations. A number of twin studies suggested wide ranges of concordance rates of between 0.23–0.86 for monozygotic twins and 0.15-0.5 for dizygotic twins7–11. These wide ranges can probably be explained in part by heterogeneity of the phenotype and phenotype definition across studies, but, as suggested by the relatively low concordance in some studies, even for identical twins, the environment is also influencing disease risk and manifestation. Parent-of-origin effects, or maternal heritability, has also been attributed to AD12, an observation which has subsequently been supported by genetic association studies (i.e., maternally transmitted alleles in the SPINK5 gene13).,
DISCOVERY OF ATOPIC DERMATITIS GENES USING TRADITIONAL APPROACHES: GENOMEWIDE LINKAGE AND CANDIDATE GENE ASSOCIATION STUDIES
Genomewide linkage studies on AD
Given the long-standing recognition of the role of heritability in the manifestation of AD and the characteristic early age of disease onset, AD is a trait highly amenable to the genomewide linkage approach for identifying novel genes. The genomewide linkage method is a family-based approach relying on a collection of affected individuals (probands) and their parents (i.e., case-parent trio design) or affected sibling pairs and their parents (and often additional family members), for which the inheritance pattern of a trait is compared with the inheritance pattern of chromosomal regions using highly polymorphic, genetic (‘microsatellite’) markers evenly spaced across all chromosomes (Figure 1, panel A). There are several advantages of genomewide linkage mapping, namely that it is ‘hypothesis-independent’, because the entire genome is scanned without regard to specific candidates. Because of the polymorphic nature of microsatellite markers, genomewide linkage mapping is cost-effective because it requires a relatively small number of markers (~350); as a result, a significant linkage peak requires a much lower correction of the p value (e.g., a lod score of 3.6, or P=2×10−5 14) compared to study designs involving thousands or tens of thousands of markers. Because linkage cannot detect genes with minimal or modest disease effect, linkage peaks that reach statistical significance are generally indicative of a locus (loci) with substantial effect on disease risk.
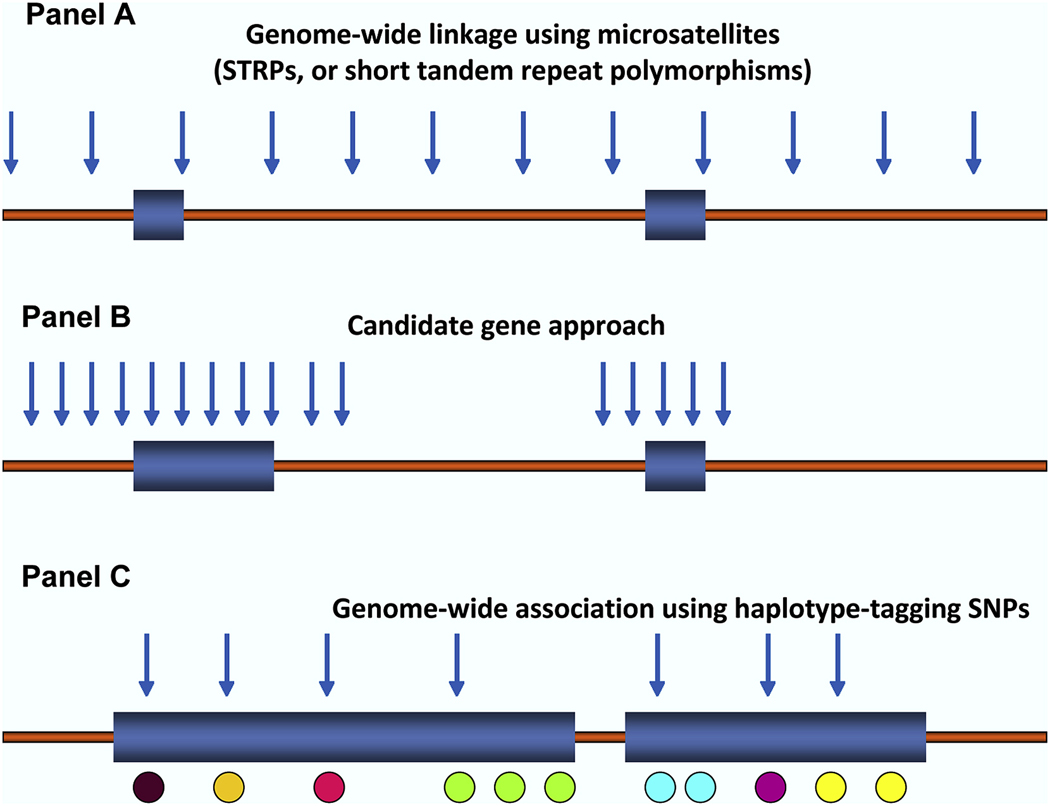
Figure 1. Methods for detecting disease susceptibility genes.
Panel A, genome-wide linkage method, relying on polymorphic microsatellites, referred to as ‘short tandem repeat polymorphisms’, or STRPs, evenly spaced across the chromosomes, typically including ~350 STRPs. Panel B, the candidate gene approach, whereby genetic markers, usually easily typed substitutions (i.e., single nucleotide polymorphisms, or SNPs) or structural variants (i.e., insertions/deletions), are selected within and flanking a candidate gene of interest. Panel C, genome-wide association studies, or GWAS, using hundreds of thousands and up to 1M SNPs to fully cover an individual’s genome, is a hypothesis-free, systematic search across the genome to identify novel associations with common diseases using a set of haplotype tagging SNPs (htSNPs), indicated as blue arrows. htSNPs represent a relatively small subset of SNPs of the potential 2.5 million SNPs in the public database (i.e., needed to uniquely identify a complete haplotype. The premise behind htSNPs is the observation that, when SNPs are in linkage disequilibrium (LD) with each other and form haplotypes, there is redundant information contained within the haplotype, because several of the SNPs will always occur together. Thus, a marker at one locus can reasonably predict a marker(s) that will occur at the linked locus nearby.
To date, there have been five genomewide linkage studies performed on AD, plus a genomewide linkage screen originally designed for asthma with analyses repeated for the AD outcome (Figure 2). All but one of these screens were performed on families of European ancestry: (i) 199 German and Scandinavian15; (ii) 148 British16; (iii) 109 Swedish17; (iv) 100 Danish18; and (v) 295 French19 families, of which 62 affected sib-pairs for AD were available for re-analysis. The non-European study was performed on 77 Japanese families selected through 111 sib-pairs with AD (287 individuals) and relied on a linkage mapping panel of 5,861 single nucleotide polymorphisms (SNPs) rather than a microsatellite panel traditionally used for linkage screens20.
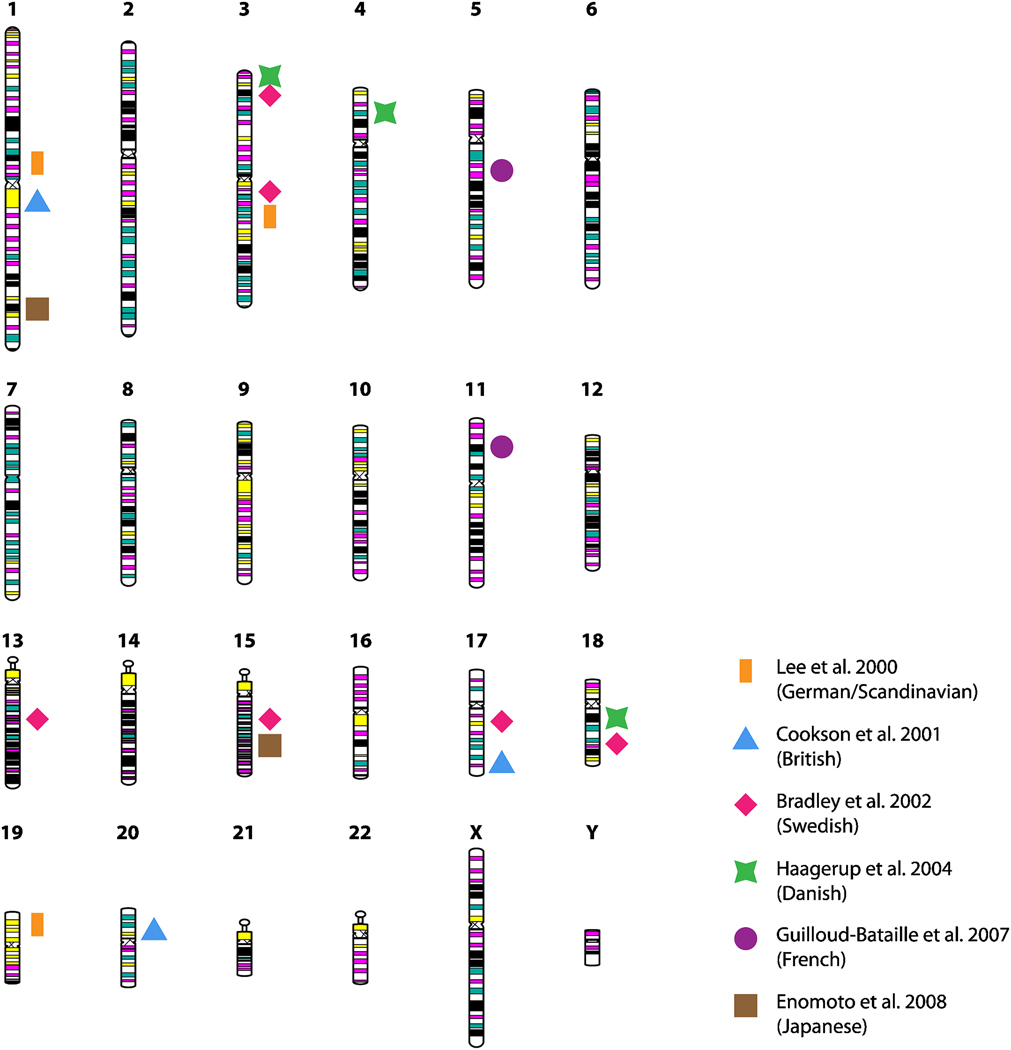
Figure 2. Summary of genome-wide linkage studies of atopic dermatitis (AD).
Representation of the 23 human chromosomes, highlighting those loci for which genome screens have identified linkage to AD. Loci are mapped to short or long chromosomal arms and color-coded according to the studies listed in the legend (Refs. 15–20).
Underscoring the heterogeneity commonly observed in complex diseases, which, in addition to genetic heterogeneity, reflects heterogeneity of non-genetic factors, including differences in family ascertainment schemes, definition of the phenotype, and analytical approaches, there has been limited overlap of signals among these genomewide linkage studies. Only the 3p24 locus has truly replicated, with significant LOD scores observed for microsatellite markers in chromosome 3p24-p22 in the Swedish17 families and chromosome 3p26-p24 in the Danish18 families. Under a more relaxed threshold of a maximum distance of 25 centimorgans (cM) between linkage peaks, replication can be considered for the chromosome 3q13–q21 locus in the German/Scandinavian15 and Swedish17 samples, and chromosome 18q11–q21 in the Danish18 and Swedish17 samples.
In the French study, the linkage screen was originally designed to study asthma and allergic rhinitis in a sample of 295 families ascertained through asthmatic probands, but analyses were repeated for the outcome ‘eczema’, and demonstrated linkage at 5q13 and 11p14. Follow-up fine-mapping of eight markers in the 11p14 locus suggested a pleiotropic effect for the three allergic diseases, AD, asthma and allergic rhinitis. More recently, the Danish team followed up on their previous evidence for linkage at the 3p, 4q and 18q loci, in addition to previous evidence for linkage at 3q, in an independent sample of 130 AD sib-pair families, and concluded the strongest evidence for linkage was to 3p34, 3q21, and 4q2221. From these follow-ups, however, conclusions are speculative at best, given that each of the regions where significant evidence of linkage has been identified contains multiple candidate genes. The best evidence for linkage in any one of these regions tends to extend over relatively large portions of the chromosome, rendering pinpointing of any specific locus – or gene – very difficult.
Although the genomewide linkage approach represented one of the most sophisticated technologies in genetic epidemiology just over a decade ago, inherent challenges in this approach including the considerable cost to follow-up genotyping of unwieldy and large chromosomal loci and the difficulties ascertaining sufficient numbers of complete families has been, in part, an impetus for pursuing alternative approaches in gene hunting. After an initial genomewide linkage analysis, positional cloning usually follows, whereby extra microsatellite markers at a density of 0.5–1.5 cM are genotyped over the linkage peaks that are suggestive or significant in the initial scan, until the precise locus contributing to linkage is identified. However, even at this level of fine mapping, with an aim of localizing a gene to a region <1 cM (1 cM.1 million base pairs), the region of peak linkage score may still include hundreds of genes. In none of the AD genomewide linkage studies illustrated in Figure 1 was a candidate gene identified using positional cloning.
Candidate genes in select pathways of AD
With the publication of initial efforts in sequencing the human genome22 (http://www.ornl.gov/sci/techresources/Human_Genome/home.shtml), the opportunity to genotype markers directly in genes of interest was greatly expanded as polymorphisms were identified in the ~20,000–25,000 genes across the 3 billion chemical base pairs that make up human DNA. Relying upon one of the simplest of these polymorphisms, single nucleotide polymorphisms (SNPs), and relatively simple structural variants, such as insertions/deletions and repeats, this advancement allowed researchers to expand genetic studies beyond linkage toward the genetic association study design (Figure 1, panel B). Because many more biallelic markers are required compared to microsatellites to detect linkage and association across the genome, a candidate gene approach was adopted whereby investigators have focused on a specific gene or set of genes believed to be causally involved in the underlying pathology of a certain disease. An advantage of the candidate gene approach is that it is not limited to families, and can be applied to case-control study designs, which possess certain advantages over family-based studies. For example, although association studies based on case-control designs are sensitive to confounding due to population stratification (i.e., ethnic/racial admixture)23, they are generally considered to be more powerful in detecting true associations once the gene has been identified24. Moreover, it is considerably easier (requires less effort, lower costs in recruitment) to amass reasonably powered groups of AD patients and healthy controls without AD than it is to collect complete case-parent trios or nuclear families.
For these reasons, there has been considerable effort in conducting association studies on AD and related phenotypes in independent populations in the post-genomewide linkage era. In a search of the public database (http://www.ncbi.nlm.nih.gov/pubmed/) through June 2009, using keywords including ‘association’, ‘atopic dermatitis’, ‘eczema’, ‘gene’, ‘polymorphism’, ‘mutation’, and ‘variant’, 111 studies were identified for which results of tests for association for AD were reported on a candidate gene (Table E1 in this article’s Online Repository at www.jacionline.org). The major outcome was limited to AD as a qualitative trait, or AD severity. A significant association was defined liberally as any association at P<0.05 and without regard to effect size or weaknesses in the study design. Limitations in this exercise include the sometimes missing information on the precise variant for which association was (or was not) demonstrated, and the unfortunate reality that negative association studies are rarely published and are therefore underrepresented in Table E1. In most instances, the only information available on genes for which association was sought but for which the result was negative are those studies for which the negative results are included in manuscripts reporting positively associated genes/variants. In other words, there are undoubtedly more genes for which associations have been tested but failed but for which the information is not available in the public arena.
It is critical to note that there are several biases in this summary. First, there is the bias of reporting associations in studies for which either Type I error (false positive) or Type II error (false negative) is possible (and in many cases, likely) due to limited power in the original study design. Other biases are related to potential study design weaknesses, such as a failure to adjust for population stratification in the case-control studies and consideration of Hardy-Weinberg proportions. There is a general assumption that, for risk variants with common allele frequencies of greater than 20%, the odds ratios (ORs) will range from 1.1–1.5, and for rarer risk allele frequencies (i.e., <0.20), the ORs can be ~3.0. Based on these assumptions, there is a requirement of at least 1,000 cases and 1,000 controls to detect ORs~1.5 with at least 80% power, although the required sample size is dependent upon multiple, additional factors. Regardless, according to the summary in Table E1, in barely two dozen of the reported studies have sample sizes reached even 250 in each group. Another important consideration is the extreme heterogeneity of the phenotype definition from study to study, wherein some studies consider early onset AD versus studies that combine pediatric populations with adult populations, variations in means for determining disease severity, and so on. Variation in the phenotype renders replication of associations especially difficult.
A critical bias in this exercise is that replication was considered at the level of the gene rather than the specific variant/mutation. Elsewhere there has been extensive discussion regarding the merit of this approach, including the comprehensive review of asthma genetics by Ober and Hoffjan23, in which the authors refer to advocacy of a gene-centric approach because the more conventional SNP-for-SNP approach is predicated on the assumption that allele frequencies and haplotype structure at a specific locus is identical in two (or more) populations, which is unlikely25. Elsewhere, the pitfalls of this “loose” replication have been cautioned26. However, a major outcome from the International Haplotype Map (HapMap) project27 is the observation that a large portion of the human genome is arranged into blocks of common polymorphisms (SNPs) in strong linkage disequilibrium (LD) with one another, and there is considerable diversity within these haplotype blocks. As much of this diversity is driven by mutation, and given our knowledge that the functional properties of protein products (e.g., candidate genes) can depend on specific combinations of multiple polymorphisms within a gene, or interactions with polymorphisms in other genes28, it is not surprising that a single-locus approach may fail to detect association a priori, much less fail to replicate across studies in different sets of populations. The recent report by Rogers and colleagues, wherein they combined SNP data from the Ober and Hoffjan review, supplemented by their own review of the asthma genetics literature, with SNP data from their genomewide association study (GWAS) on over 1,000 asthmatic children and family members in the CAMP study and >500,000 SNPs, underscores the unlikely success in replication at the SNP level, as the group only identified 10 significantly associated SNPs in six genes that were on the commercial SNP chip and that overlapped with 160 SNPs in 39 genes previously associated with asthma in the literature29.
Finally, another area of potential controversy has to do with common versus rare variants. A perspective on the scientific value of GWAS30 suggested that (assuming currently available commercial chips capture the bulk of common genetic variation) SNPs with large effects may have already been discovered (implying there is no need for additional genome wide scans), and the focus should therefore shift towards detailed search for rare variants. Although the application of both conventional candidate gene studies as well as GWAS has lead to the discovery of hundreds of loci conferring disease risk of common diseases, it has been noted that the risk variants identified by GWAS in particular explain only a small fraction of the disease-specific heritability. One explanation that is beginning to emerge is that much of the remaining heritable disease risk is associated with rare variants31 (usually defined as those with a frequency of less than 5%−1% or lower). For example, rare alleles of at least 10 loci contribute to risk for breast cancer, and more than 40 loci underlie risk of type 1 diabetes. Importantly, in contrast to common variants, rare nonsynonymous variants in the human genome may be deleterious and therefore of significance because they influence protein function and/or phenotypic variation. Most of these variants are probably only mildly deleterious, segregate at low frequencies, and not pathogenic. However, a subset of these variants have more modest effects and are rare, but could collectively explain a substantial fraction of the heritably variability of common disease phenotypes.
While rare coding variants may have a greater functional impact than common variants, their analysis must consider the low frequency of any variant since it will reduce the power to infer statistical associations, and therefore require very large populations of cases and controls or affected families. This complication can be overcome by evaluating the collective frequency of rare, nonsynonymous variants within one or more genes, or for a pathway(s) and the functional impact of the discovered variations. As described further below, however, with the exception of studies focused on mutations in the gene encoding filaggrin (FLG), few if any other studies in AD have specifically focused on rare variants nor have designed studies of sufficient power to be able to detect rare variants.
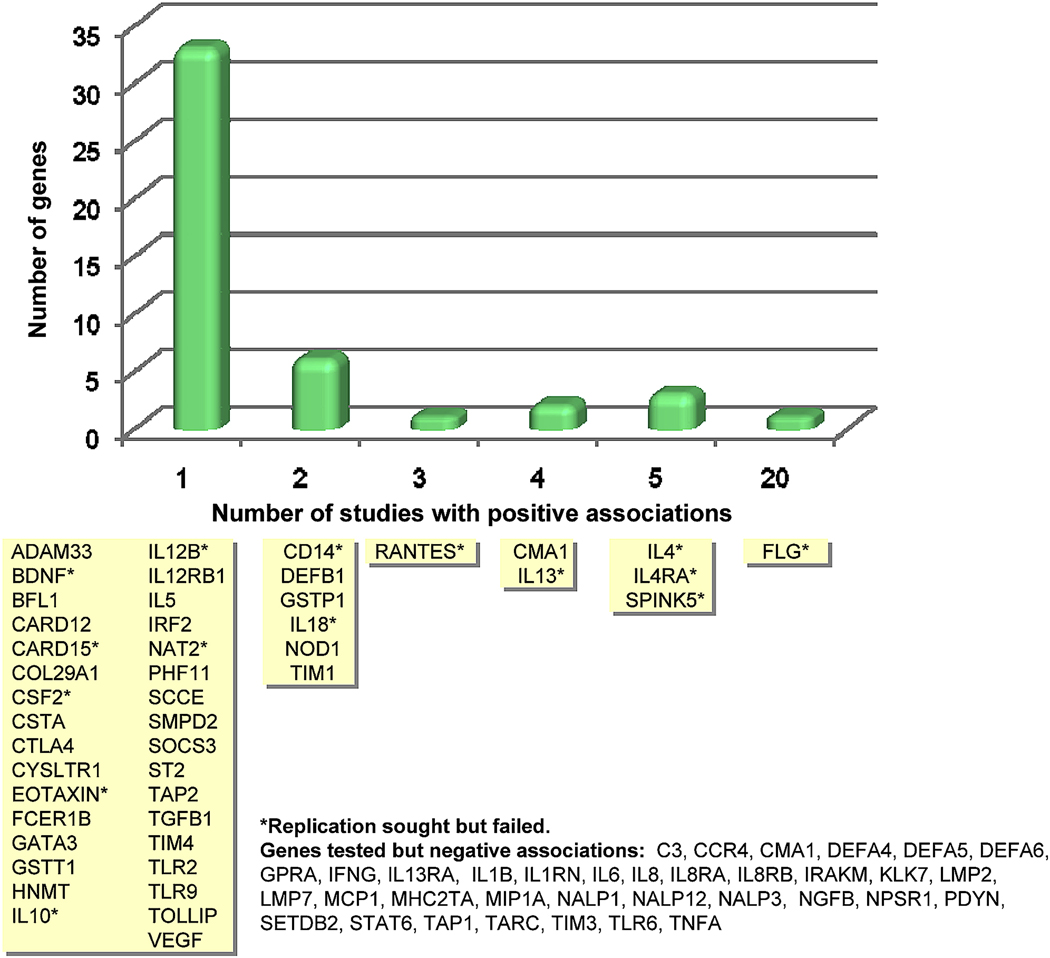
Despite the biases and potential pitfalls described above, several interesting conclusions can be inferred from this comprehensive review of the literature on the genetics of AD. In the 14 years since the first published report on an association between a genetic variant and AD, more than a third of the studies have been reported in the past two and a half years, and already in the first half of 2009, more reports have been published than in all of 2008. From these 111 published studies, there are reports on 81 genes, of which more than half (46 genes) had at least one positive association study reported (Figure 3). Of these 46 genes, 15 studies failed to replicate associations, and 13 were positively associated in at least one other independent study. One of these genes, the gene encoding filaggrin (FLG), has been associated with AD in 20 different reports (details below). There are 35 additional genes studied for which there has been no evidence for a positive association to date.
Figure 3. Genes associated with atopic dermatitis in at least one published study.
Genes are grouped according to how many positive association studies have been reported (see Table EI in this article’s Online Repository at www.jacionline.org for a complete summary of 111 published studies). The Y-axis indicates the number of genes (corresponding to the yellow boxes) for each time positive association was reported.
Adaptive and Innate Immune Response Genes
The well-established co-morbidities of the other two allergic diseases that constitute the atopic triad (i.e., asthma, allergic rhinitis), in addition to sensitization to common aeroallergens that is commonly observed in patients with AD, has guided researchers toward selection of candidate genes that fall within the broad category of dysregulation of the adaptive and innate immune response and a heightened IgE-mediated, systemic Th2 response, plus a combined Th1 and Th2 response in the skin. This selection bias is evident in Table E1.
For complex diseases such as AD that involve a dense network of immune response proteins, it is anticipated that many genes will be involved and multiple genetic variants will contribute to the alteration of gene function and expression. Because the effect of a single gene/polymorphism in a complex disease such as AD is anticipated to be relatively modest, it can be assumed that variants in multiple genes will cooperate in an additive or synergistic manner to impact disease risk, a phenomenon referred to as ‘epistasis’. A major difficulty in testing for epistasis is power: not only will fewer individuals in a sample possess both polymorphisms (or a set of polymorphisms) associated with risk of disease compared to the number of individuals with only one of the polymorphisms, but the correction factor for additional (multiple) comparisons (i.e., tests for association) is quite large. Consider the example of 100,000 genetic markers, wherein there are a total of 5×10+9 two-locus combinations, requiring a (Bonferroni) correction factor of P=1×10−11 for a GWAS significance level of 0.0532.
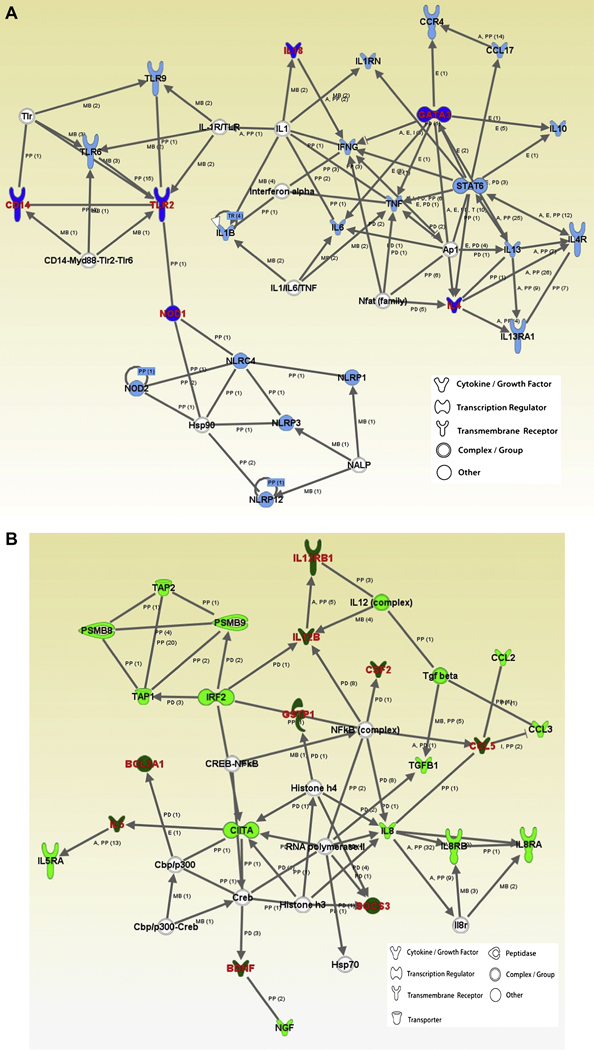
One approach toward characterizing potential gene-gene interactions and systematically evaluating the role of candidate genes/polymorphisms in AD susceptibility for which there is compelling evidence for association is to implement the program Ingenuity Pathways Analysis (Ingenuity Systems: www.analysis.ingenuity.com). The Ingenuity Pathways knowledge base is a web-based entry tool developed by Ingenuity Systems Inc. to characterize genes according to the pre-defined canonical pathway(s) into which they fit, and also to investigate the extent to which genes are in shared networks and may cooperate in a synergistic or an additive manner to impact risk of disease. As a proof of concept, we evaluated the 81 genes summarized in Figure 3 using the Ingenuity Pathway Analysis. Slightly more than half (N= 48) of the 81 genes studied to date clustered into two major networks, both of which are associated with immune dysregulation, specifically the pathway associated with antigen presentation and cell-mediated and humoral immune response, and the pathway associated with cell signaling and interaction, cellular movement, and hematological system development and function. Six genes (CD14, GATA3, IL4, IL18, NOD1, TLR2) which have previously been significantly associated with AD were clustered into the antigen presentation and immune response pathway, and nine previously associated genes (BCL2A1, BDNF, CCL5, CSF2, GSTP1, IL5, IL12B, IL12RB1, SOCS3) were clustered into the cell signaling/movement pathway. Although the studies for which these candidate genes were evaluated did not specifically test for gene-gene interaction, this interrogation of the potential for interaction serves as an example of the power of this approach in selecting optimal candidates for genetic association studies.
In addition to immune dysregulation manifested as IgE-mediated sensitization to numerous allergens, AD is also characterized as a common, chronic pruritic, inflammatory skin disease complicated by recurrent bacterial and viral skin infections (i.e., Staphylococcus aureus and herpes simplex virus)33, 34. More seriously, patients with AD are at greater risk for developing severe and generalized viral infections caused by herpes simplex virus (e.g., eczema herpeticum), molluscum contagiosum virus (e.g., eczema molluscatum), and eczema vaccinatum, which occurs after exposure to the smallpox vaccine35.
Increased susceptibility to infections and cutaneous colonization implicates several of the immune function genes listed in panel A of Figure 4, specifically those genes associated with a dysfunctional host defense – or innate immune – response. These candidates include the pattern-recognition receptors (PRRs) type I transmembrane, toll-like receptors (TLRs)36, the nucleotide-binding oligomerization domain (NOD)-leucine rich containing protein family (NOD1, NOD2)37, 38, and CD1439–41. Antimicrobial peptides, including S100 proteins, human defensin-α and –β, and sphingosine exert potent antimicrobial activity by directly killing bacteria, fungi, and certain viruses42. Natural killer (NK) cells, a critically important population of lymphocytes for innate immune responses against virus infection43, are dependent upon transcription factors such as IFN regulatory factor-2 (IRF-2) for efficient cell development (for an extensive review on innate immunity in AD, see Refs. 44 and 45). Genetic association studies on AD to date in fact support a number of these candidates (i.e., TLR2, NOD1, NOD2, CD14, DEFB1; Figure 3). In addition to the direct effect of genetic modifications in innate immune response molecules and their role in susceptibility to infection, the attenuation of upregulation of the normal antimicrobial response to bacterial and viral stimuli because of an overabundance of Th2 cytokines in the skin appears to be especially relevant in AD46, 47. For example, it has been demonstrated that elevated Th2 cytokine levels can inhibit mobilization of potent innate immunity molecules, such as human beta-defensin-3, in epidermal keratinocytes48. Alternatively, persistent S. aureus infections can also mediate inflammatory cascades by staphylococcal toxins acting in a superantigen-driven fashion to activate T cells49 or by induction of a state of glucocorticoid resistance50. This observation further underscores the potential relevance of gene-gene interactions, as illustrated in panel A of Figure 4.
Figure 4. Networks revealed through the Ingenuity Pathways Analysis based on 81 AD genes established in genetic association studies.
48 genes clustered into two major networks associated with immune dysregulation. Panel A, antigen presentation and cell-mediated and humoral immune response pathway. Significantly associated genes are highlighted in dark blue. Panel B, cell signaling and interaction, cellular movement, and hematological system development and function pathway. Significantly associated genes are highlighted in dark green. For both panels, genes or gene products are represented as nodes (shapes; see legend) that represent the functional class of the gene product. Relationship between genes is presented as solid lines with arrows (direct activation) and arrows point to the element on which an action is performed. All relationships (or lines) are supported by at least 1 reference (numbers of references in parentheses). Annotation of relationship (labels) between the nodes: A – activation, E - expression, I – inhibition, MB - group/complex membership, PD - Protein-DNA binding, PP - protein-protein binding.
Notably, the candidates described above and summarized in Figure 4 as well as Table E1 are not all unique to AD; in fact, many AD candidate genes overlap with not simply other atopic phenotypes (e.g., asthma, allergic rhinitis), but also other diseases of inflammation and immune dysfunction. The common disease/common variant hypothesis41 has been put forth as one explanation for why many complex diseases are so common and why disease-associated variants occur at such high frequency in the population. The ‘common variant/multiple disease’ hypothesis, which is an extension of the common disease/common variant hypothesis, suggests that certain disease genes may not be disease specific, and may contribute to related clinical phenotypes51. One possibility is that the functional effects of certain alleles manifest in multiple disorders, presumably because they are involved in basic underlying immune regulatory pathways. Multiple examples of genetic association of the same gene (or allele) to diverse but related disorders abound (see the Genetic Association Database at http://geneticassociationdb.nih.gov). For example, in addition to AD, genetic associations have been observed DEFB1 for asthma52, COPD53, and infectious disease including HIV54 and sepsis55. NOD2 polymorphisms, in addition to AD, have also been associated with Crohn’s Disease56 and sarcoidosis57. Associations with RANTES polymorphisms are especially diverse, including phenotypes associated with immune response, infection, reproduction, and metabolic disorders (http://geneticassociationdb.nih.gov/). Given the prominent role that certain pathways, such as host defense, play in susceptibility to AD, it is likely that many more co-associations will be observed in the near future.
Skin Barrier Dysfunction Genes
It is increasingly appreciated that both genetic and environmental factors that affect skin barrier function contribute to AD susceptibility58, and that barrier dysfunction is an essential feature of AD and allergic diseases in general59–63. A disrupted barrier would allow penetration of microbes and allergens and other environmental insults such as toxins, irritants and pollutants with consequences including inflammation, allergen sensitization and bacterial colonization. This may explain why 55%–90% of patients with AD are colonized with S. aureus compared to only 5% of individuals without AD64–68. Although the epidermis functions as the primary defense to the external environment, considerable barrier function is regulated by the stratum corneum (SC) and, below the SC, tight junctions (TJ), which reside at the level of the stratum granulosum. When the SC is compromised, either by reduced levels of SC lipids29–31, mechanical trauma resulting from extensive scratching that is precipitated by intensive itch (the hallmark of AD), or as a result of genetic defects in SC proteins (i.e., filaggrin), tight junctions are the next line of defense. Currently there is considerable interest in the more than 40 proteins that comprise TJ, which include the claudin family members, occludin, cingulin, tricellin, and the cytoplasmic plaque proteins69 (which bind to actin and myosin70), and their role in human disease in general71, and specifically AD.
As described earlier, linkage screens performed on AD to date have not elucidated specific candidate genes per se, but they have implicated loci harboring clusters of genes associated with skin barrier dysfunction. Specifically, one of the earliest screens indicated linkage at the epidermal differentiation complex (EDC) locus on chromosome 1q2116, which contains a very large and diverse family of genes associated with skin barrier dysfunction, including loricrin (LOR), involucrin (IVL), members of the S100 gene family, a large group of the late cornified envelope gene family, many of the small proline-rich proteins, peptidoglycan recognition proteins (i.e., PGLYRP3, PGLYRP4) and, most notably filaggrin (FLG)72. Linkage has also been reported in 17q2117, where the gene encoding one of the keratins (KRT16) is localized.
Association studies on genes related to the EDC cluster and other barrier dysfunction candidates have, to date, been largely restricted to the gene encoding filaggrin (FLG), also known as filament-aggregating protein, and within FLG, most associations have been limited to two null mutations (R501X and 2282del4). In fact, FLG is the most consistently associated gene with risk of AD73; as illustrated in Figure 3, by mid-2009, there were 20 positive reports on genetic associations between FLG mutations and AD. The gene encoding human filaggrin was first cloned in 1989, when it was found to contain numerous tandem filaggrin repeats, localized to chromosome 1q21, and, because of its tight regulation at the transcriptional level in terminally differentiating epidermis, it was postulated to be an important candidate for disorders of keratinization74. It was subsequently evaluated for its function in the formation of the stratum corneum, and found to be a critical protein involved in epidermal differentiation and in maintaining barrier function75. McLean and colleagues developed long-range PCR conditions for a 12-kb genomic fragment covering exon 3 of FLG, and identified a homozygous nonsense mutation (R501X) near the start of repeat 1 and a second mutation (2282del4) that similarly stops protein translation within the first filaggrin repeat in three patients with ichthyosis vulgaris76. They demonstrated that these relatively rare mutations (a combined allele frequency of ~4% in the European population studied) are semidominant, with heterozygotes exhibiting mild disease with incomplete penetrance. Shortly thereafter, the same group showed that these two loss-of-function variants were associated with AD77, and that they are ancestral European variants carried on conserved haplotypes 78.
Full sequencing of the FLG gene by the same team has revealed multiple, additional polymorphisms with varying frequency across ethnic groups78; however, with a combined allele frequency among AD patients of 18% and 48% for the R501X and 2282del4 mutations, respectively, the two null mutations represent the strongest and most compelling genetic risk factors for AD. In the largest meta-analysis performed so far on the R501X and 2282del4 mutations, Rodriguez and colleagues79 analyzed data from 24 independent studies, which included 6,448 cases, 26,787 controls, and 1,993 families (all selected for AD), and determined that the effect size for risk of eczema due to the two FLG null mutations is not dissimilar to previous reports, at an odds ratio of just over three.
The FLG mutations have also been consistently associated with risk of other atopic traits, including asthma, hay fever, rhinoconjunctivitis, and allergen-specific IgE79–86. Recently, Gao and colleagues (unpublished data) evaluated whether FLG polymorphisms contribute to the serious complication of AD resulting from disseminated cutaneous HSV infections, eczema herpeticum, and determined that the frequency of the R501X mutation was three times higher (24% vs. 8%, respectively) and the relative risk for disease nearly double for eczema herpeticum compared to AD without eczema herpeticum (odds ratio [OR]=11.8 vs. 6.2; P=0.0008). The authors speculate that the relationship between FLG null mutations and disease is most likely related to an increased propensity to disseminated viral skin infection resulting from skin barrier dysfunction, rather than disease severity per se, and provide the first insight into the genetic underpinnings of AD complications such as viral dissemination.
New approaches on the horizon: The genomewide approach
Genomewide association studies, or GWAS
A major limitation in the candidate gene approach is that selection of candidates is often based on limited knowledge87 and, moreover, each potentially causal variant at each candidate gene can only make a modest contribution to overall heritability. Data from the HapMap Project, combined with more accurate approaches in selecting tagging markers sufficiently dense to capture most of the common variation in the human genome (Figure 1, panel C), have recently allowed genomewide association studies (GWAS) to replace candidate gene studies as an unbiased approach to search for genes controlling risk to complex diseases. At the time of this review, results have been published on one GWAS for AD, in which investigators in Germany genotyped a set of AD cases and controls and an independent set of 270 nuclear families on the Affymetrix Human mapping 500K and 5.0 arrays (resulting in 342,303 successfully typed markers)88. Although none of the markers analyzed were significantly associated with AD at the genomewide significance level (P<1.46×10−7), the group genotyped 54 SNPs associated with AD at a modest P-value in an independent German group and observed replication in markers in chromosomal regions 1q21, 9p21 and 11q13, with further replication in an additional European group for a marker (rs7927894) in an intergenic region near C11orf30 on chromosome 11q13.5, which is the same marker that has been associated with Crohn’s Disease89. An additional European-based GWAS is in the replication phase. It will be of interest to determine the extent to which the Esparza-Gordillo et al. findings will be replicated, or not.
High-throughput gene profiling
Expression profiling of all known genes in the human genome is an ideal strategy for characterizing disease mechanisms and defining the transcriptome of complex diseases such as AD. Indeed, genomewide microarray technology has the potential to identify molecular signatures of clinical disease impossible to identify using a gene-by-gene approach, and there are many examples whereby this technology can be highly predictive of clinical outcome. Although relatively few genomewide expression studies have been performed for AD, which is likely due to the difficulties in ascertaining sufficient numbers of selected cell types of the epidermis and other relevant tissue samples, limitations related to the size of biopsied tissue, and challenges in ensuring collection of representative samples, it is anticipated that integration of this technology into gene discovery for AD will increase, especially as the cost for performing genomewide microarray continues to drop.
Sugiura and colleagues90 performed high-throughput expression profiling of biopsies from skin lesions of AD patients compared to healthy controls, and observed that several of the most significantly differentially expressed genes (i.e., S100A8 and S100A7, upregulated; loricrin and filaggrin, downregulated) were epidermal differentiation genes localized in chromosome 1q21, a region previously linked with AD (note: results were similar in comparisons between affected and unaffected skin among the AD patients). Similar findings have been observed by Beck and colleagues, who compared nonlesional skin of extrinsic AD subjects to the skin of nonatopic healthy controls (personal communication; see Figure 5B). The Sugiura group also observed that keratin 16, localized on chromosome 17q21, another linkage hotspot, was upregulated. In addition to validating genes, high-throughput expression studies have led to novel discoveries. One example is the work by Lu and colleagues91, where they relied upon primary cultured keratinocytes from AD patients and healthy controls to identify a number of novel candidates. Significant findings included two extracellular matrix-associated factors, MMP1 and MMP10, for which ELISA studies of intrinsic AD patients’ serum showed that both proteins were up-regulated nearly two-fold compared to healthy individuals and extrinsic AD subjects. In a differentiated keratinocyte model, filaggrin 2 (or ifapsoriasin), also localized in the EDC, was identified, as well as three novel lipase genes, suggesting that these genes may also play a key role in the skin barrier and are worthy of further study92. Thus, high-throughput expression profiling has already proved useful in supporting the hypothesis that defects in epidermal genes play a critical role in the development of AD, and it is also a tool for validating genetic findings and gene discovery.
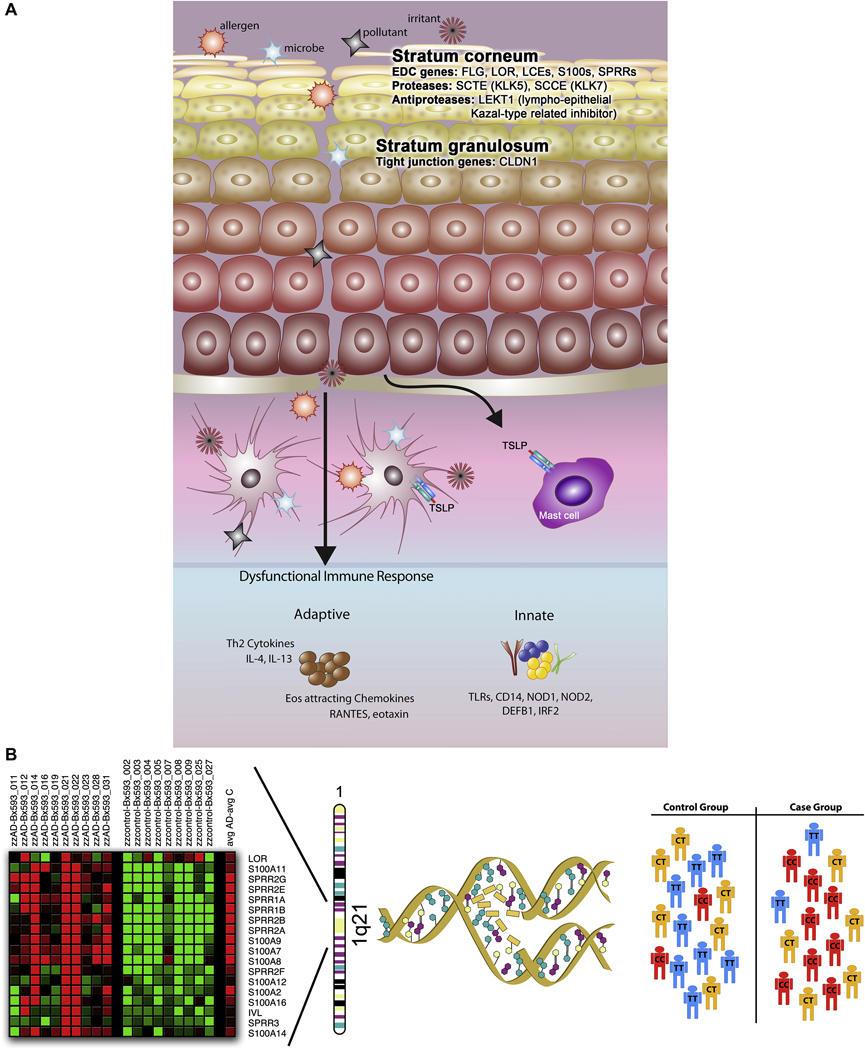
Figure 5. Genetic epidemiological approaches toward identification of genes conferring AD susceptibility.
In this summary of approaches used to date to identify candidate genes for AD, the first consideration is the complex pathology of AD (panel A), which includes defects and damage at the epithelial barrier, and penetration of allergens, microbes, pollutants and irritants into the epidermis and dermis, ultimately interacting with antigen presentation cells (i.e., Langerhans cells and dermal dendritic cells). The ‘brick wall-like’ structure of the stratum corneum is compromised, possibly by defects in genes residing in the epidermal differentiation complex (EDC), including FLG, LOR, LCEs, S100s, SPRRs, SCTE, and SCCE. Additional candidates include tight junctions (TJ), proteins that constitute the “gate” to the passage of water, ions and solutes through the paracellular pathway in the stratum granulosum (i.e., CLDN1). A defective innate immune response (involving, for example, the TLRs, CD14, NOD1, NOD2, DEFB1, and IRF2) may contribute to a heightened IgE-mediated, systemic Th2 response. Combining a candidate gene approach with robust, genomewide gene expression profiling has the potential of both elucidating novel candidates and validating suspected candidates, as illustrated in panel B, in which genes in the EDC on chromosome 1q21 are significantly differentially expressed in a human skin biopsies taken from AD patients compared to healthy nonatopic controls (courtesy L.A. Beck). Candidate genes are selected, and substitutions (i.e., single nucleotide polymorphisms, or SNPs) and simple structural variants (i.e., insertions/deletions, repeats) are genotyped in large populations selected for AD and tests for association are performed.
Confounders and complexities
It’s not all about the genes: the role of the environment
In the same year that Watson and Crick reported their structure of the DNA molecule in the journal Nature93, Senior Registrar at the Manchester and Salford Hospital for Skin Diseases, Dr. J.K. Morgan, made the following observation: “In theory, in every case in which there is an eczematous reaction in the skin, there are two prime factors involved. The first, and the more important, is a constitutional or internal factor, relating to the individual himself. The second in an external factor. It is upon the relative balance and interplay between these two that the appellation of the disease is based”94. In the years of heritability studies that followed (described above), the consistent observation that concordance rates for AD among monozygotic twins raised together are higher than among dizygotic twins supported the role of genetic etiology, but the relatively low concordance rates in both groups also supported the earliest suppositions that differences in exposure to certain environmental triggers may account for a considerable proportion of disease expression.
The challenge in the genetic epidemiology of AD in terms of interrogating gene-environment interactions is precisely which environmental factors should be considered. For example, at the core of the innate immune response are the ubiquitous fragments of bacterial LPS, or endotoxin, and many studies focusing on endotoxin exposure early in life suggest a ‘protective’ effect in the development of allergic disease in general95, 96 by skewing the Th profile toward Th1, as purported by the ‘Hygiene Hypothesis’97. Interestingly, there is some support for the role of the Hygiene Hypothesis in susceptibility to AD98. Alternatively, the impact of exogenous substances, such as irritants (i.e., soap and detergents), allergens (i.e., exogenous proteases derived from house dust mites), and drugs (i.e., topical corticosteroids) on AD patients with genetic alterations in skin barrier genes has also been considered58.
Very few of the genetic association studies summarized in Table E1 (Online Repository at www.jacionline.org) have considered evidence of association between genetic polymorphisms in the context of exposure to certain environmental factors. Perhaps the best example is the study by Bisgaard and colleagues99, in which it was hypothesized that a compromised skin barrier among individuals with AD who are filaggrin-deficient may enhance the impact of exposure to certain aeroallergens, such as house dust mite and pet allergen. The group confirmed previous observations that the risk of developing eczema was considerably higher among children with the FLG mutations (hazard ratio 100=2.26, P=0.0005), but that the risk increased considerably if children were exposed to cat allergen at birth (HR=11.11, P<0.0001). Important considerations in future, similar studies will be the ability to detect such interactions for variants with smaller effect sizes, the temporal relationship between environmental exposure and risk of disease (i.e., perinatal exposure versus exposure later in childhood), and the generalizability of such findings across populations.
Ethnic diversity and underrepresentation
As described previously, failure to replicate associations between genetic markers and a complex trait such as AD in independent populations can be due to several factors, including chance, misclassification of the phenotype, environmental heterogeneity, inadequate sample sizes, and population stratification101–103. An important factor contributing to failure to replicate associations is also related to population diversity. For example, it is possible that certain genetic markers may contribute to disease risk in one particular (i.e., ethnic, racial) population but not in others, either because of differences in frequencies of the risk allele(s), or because of specific gene-gene interactions. It is difficult to evaluate the impact of ethnicity on genetic associations of AD, however, because there is relatively little diversity in the populations that have been studied thus far. As illustrated in Table E1 (Online Repository at www.jacionline.org), the overwhelming majority (N=70) of association studies have been performed in populations of European descent, followed by 42 studies performed in Asian populations. A single study was performed in a Mexican cohort, and none of the reported studies have been performed in groups considered underserved minorities, such as African Americans. Sadly, this statistic does not reflect the actual prevalence of AD, where African Americans and Asian/Pacific Islanders reportedly suffer more from AD than U.S. whites104.
Perhaps the best example of a candidate gene for which ethnicity likely influences the extent to which a polymorphism confers risk is FLG. Palmer and colleagues77 observed differences in the R501X and 2282del4 FLG null mutation frequencies in diverse cohorts and suggested that different populations will have different FLG mutation profiles. Indeed, this group and others have demonstrated that, in populations where the R501X and 2282del4 mutations are not present, other mutations are prevalent and confer risk of AD, for example, the 3321delA and S2554X mutations among Japanese patients105. In unpublished data, our group observed a complete absence of the R501X mutation and very low frequency (1%) of the 2282del4 mutation among healthy African Americans, compared to a frequency of 9% of the R501X mutation and 0% of the 2282del4 mutation among AD patients (unpublished data). The low frequency and even absence of this mutation is not novel; elsewhere the prevalence of the R501X mutation among individuals without AD has ranged from 0.8% to 3.0% among European populations, and has been found to be absent in southern European (i.e., Italian106) and Asian77, 105 groups. In the only summary data available on frequency of this mutation in African populations, it was absent in a cohort of 124 North Africans77. Collectively, the distribution of these alleles suggests a latitude-dependent distribution with a decreasing north-south gradient of frequency, and suggests that polymorphisms other than the relatively common R501X and 2282del4 mutations may be more important in non-Northern European groups.
Summary
To recapitulate, AD is a chronic inflammatory disease of the skin characterized by dysregulation of the adaptive and innate immune response and a heightened IgE-mediated, systemic Th2 response. Extreme Th2 polarization and primary defects in the innate immune response, including epithelial barrier defects, in conjunction with mechanical damage to the epidermis as a consequence of the intense pruritus that is the hallmark feature of AD, likely contribute to the more severe sequelae including chronic bacterial colonization (i.e., S. aureus infection) and viral dissemination (i.e., eczema herpeticum). This scenario can be summarized in Figure 5, whereby the damaged epidermal surface is penetrated by a host of exogenous substances, including allergens, irritants, microbes, pollutants, and even topical drugs. The ‘brick wall-like’ structure of the stratum corneum (SC), which normally creates a barrier that maintains water within the body and prevents the entrance of pathogens and allergens, is further compromised. The epidermal differentiation complex (EDC), the DNA region where a large number of genes encoding many of the cornified cell envelope precursor proteins, small proline-rich proteins, members of the S100 family, and intermediate filament-associated protein precursors (i.e., profilaggrin), are localized, is an important target of candidate genes associated with barrier dysfunction at the level of the SC. The last line of defense is the stratum granulosum, containing TJs, proteins that constitute the “gate” to the passage of water, ions and solutes through the paracellular pathway. Systemically, a dysfunctional immune response resulting in an imbalanced innate and adaptive milieu further aggravates the system. Early genomewide linkage studies, association studies, and high-throughput expression profiling studies have supported the role of skin barrier dysfunction candidate genes in conjunction with innate and adaptive immune response genes. A comprehensive evaluation of all candidate gene studies published to date on AD illustrates the importance of both sets of genes, and a pathway analysis of the genes studied so far supports a more thorough approach toward gene-gene and gene-environment interactions.
There are considerable challenges in the field: expanded analyses of skin barrier dysfunction genes, to include not only those residing in the stratum corneum, but also tight junction (TJ) genes at the level of the stratum granulosum; an expanded focus on rare, in addition to, common variants; expansion of population studies to include more ethnically diverse groups that are adequately powered; and a better integration of higher throughput technology in addition to analyses that consider the interactive effects of common environmental factors. Each of these efforts will require greatly expanded sample sizes of carefully phenotyped patients and rigorous statistical approaches. None of these goals are achievable in the absence of a multidisciplinary approach, which will require the equal contributions of expert clinicians, geneticists, statistical analysts, and molecular biologists.
What do we know?
It has long been recognized that there is a heritable component to the development of atopic dermatitis (AD).
Five genomewide linkage studies have been performed on AD, plus a genomewide linkage screen originally designed for asthma with analyses repeated for the AD outcome; however, there has been limited overlap of signals across studies with only the 3p24 locus replicating.
For the majority of candidate gene studies on AD to date, the focus has been on single nucleotide polymorphisms (SNPs), which are relatively simple structural variants, and include substitutions, insertions/deletions and repeats.
In a search of the public database for references through June 2009, there were 111 studies reporting on genetic association for 81 genes, of which more than half (46 genes) had at least one positive association.
Mutations in the gene encoding filaggrin (FLG) have been associated with AD in more than 20 different reports.
Using a pathways analysis approach, slightly more than half (N= 48) of the 81 genes studied to date for AD cluster into two major networks: (i) antigen presentation and cell-mediated and humoral immune response and (ii) cell signaling and interaction, cellular movement, and hematological system development and function.
There is increasing evidence that genes associated with skin barrier dysfunction, especially genes that reside in the epidermal differentiation complex (EDC) locus on chromosome 1q21, are associated with risk of AD.
The first genomewide association study (GWAS) on AD has been reported, and although none of the associations approached a genomewide significance threshold, replication of modestly associated markers was observed in independent populations for a marker near C11orf30 on chromosome 11q13.5, the same marker associated with Crohn’s Disease.
Expression profiling of all known genes in the human genome has been useful in validating several candidate genes, including several candidates in the EDC.
What is still needed?
Interpretation of results from candidate gene studies of AD is difficult because of a variety of potential shortcomings, including the bias of journals in only reporting positive associations, power limitations in the original study design, and a failure to adjust for population stratification (i.e., admixture of different ethnic/racial groups) in the case-control studies and consideration of Hardy-Weinberg proportions.
It is generally believed that most polymorphisms will confer a relatively small effect on risk of disease; however, most published studies to date on AD are insufficiently powered to detect an Odds Ratio (OR)~1.5.
It is increasingly believed that rare variants may contribute considerably to risk of complex, common diseases such as AD, but very few studies to date have focused on the role of rare variants.
Although environmental factors are believed to contribute greatly to the manifestation of AD, there is a dearth of information on the role of gene by environment interactions.
Some polymorphisms may contribute to disease risk in one particular ethnic/racial group but not in others, but very few genetic association studies in non-white populations have been performed.
Supplementary Material
Acknowledgements
The author wishes to thank Drs. Li Gao, Peisong Gao, and Candelaria Vergara, Nicholas Rafaels, and Pat Oldewurtel for technical assistance, and Mr. Boyd Jacobson for his artistic contributions, as well as Drs. Donald Leung and Stephan Weidinger for invaluable discussions and comments. A special thanks to Dr. Lisa A. Beck, who shared critical preliminary data and contributed to important discussions. The author gratefully acknowledges the contributions of the Atopic Dermatitis Vaccinia Network (ADVN) in generating much of the data used in this review.
Funding Sources: This study was supported by National Institute of Health (NIAID: HSN266200400029C).
ABBREVIATIONS
- AD
Atopic Dermatitis
- AIMs
Ancestry Informative Markers
- CDCV
Common Disease/Common Variant
- cM
Centimorgans
- EDC
Epidermal Differentiation Complex
- FLG
Filaggrin
- GK
Granular Keratinocyte
- GWAS
Genomewide Association Study
- HapMap
Haplotype Map
- IRF-2
IFN Regulatory Factor-2
- IVL
Involucrin
- LD
Linkage Disequilibrium
- LOR
Loricrin
- LPS
Lipopolysaccharide
- MAFs
Minor Allele Frequencies
- NK
Natural Killer
- NOD
Nucleotide-Binding Oligomerization Domain
- OR
Odds Ratio
- PRRs
Pattern-Recognition Receptors
- SC
Stratum Corneum
- SNPs
Single Nucleotide Polymorphisms
- TJ
Tight Junctions
- TLRs
Toll-Like Receptors
Barnes Clinical Glossary
- CD14
CD14 is the pattern recognition receptor for lipopolysaccharide (LPS) and is present on monocytes, macrophages, and neutrophils; genetic polymorphisms in the CD14 gene have been linked to gene-environment interactions including dog ownership, house dust mite exposure, and tobacco exposure in asthmatic patients.
- Claudin, Occludin
Claudins and occludins are members of the tight junctions that are important for epithelial barrier integrity. Claudin-1 gene deficient mice die shortly after birth and have significant transepithelial water loss.
- Expression Profiling
Expression profiling can assess the transcriptional activation of large sets of genes using microarrays that identify the activity of target genes in control versus diseased populations; sequence based expression profiling can sequence any active gene rather than a defined target set of genes on a microarray.
- Filaggrin
A skin matrix protein that promotes keratin aggregation. Mutations in filaggrin have been associated with icthyosis, eczema, and asthma.
- GWAS
Genome wide association studies are used to find candidate genes linked to a disease of interest using diseased and control patients. The screen uses bioinformatics to find associations between the disease and a haplotype block based on single nucleotide polymorphisms. Once a block is found, a candidate gene of interest that resides in the block can be studied to see associations with the disease of interest. A recent asthma GWAS found a new candidate gene ORMD3.
- Haplotype Blocks
A group of single nucleotide polymorphisms that is statistically associated such that the identification of a few alleles will define all of the polymorphisms in a given region of the chromosome.
- Human beta defensins
Innate antimicrobial peptides that are important as the first line of host defense, HBDs are decreased in the skin of patients with eczema and appear to predispose to cutaneous superinfections
- Involucrin, Loricin
Loricin and involucrin (along with profilaggrin) are found in basophilic keratohyalin granules of granular keratinocytes. Release of granule contents into the intracellular space between the granular and cornified layers of the skin are essential for protection from transepidermal water loss.
- Linkage Disequilibrium
Linkage disequilibrium refers to the nonrandom association of two or more gene loci. The degree of linkage disequilibrium measures the frequency in the combinations of alleles in a population.
- Pattern Recognition Receptors
Families of receptors in the innate immune system that recognize pathogen associated molecular patterns and danger associated molecular patterns including peptidoglycan, RNA, muramyl dipeptide, and amyloid-β-peptide.
- NOD
Nucleotide binding and oligomerization (NOD)-like receptor family protein members are members of the pattern recognition receptor family that includes Toll like receptors (TLRs). Mutations in NOD-like receptor genes have been implicated in a number of disease including Muckle-Wells syndrome, Crohn’s disease, and vitiligo.
- S100
A family of proteins that contain calcium binding activity and regulate the cell growth, cycling, and differentiation. There are 20 S100 protein family members, 14 of which reside on chr1q21
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Smith WD. Hippocrates. Cambridge, MA: Harvard University Press; 1994. [Google Scholar]
- 2.Besnier E. Premier note et observations preliminaire pour service d-introduction a letute des prurigos diathesiques. Ann Dermatol Syphil. 1892;3:634–638. [Google Scholar]
- 3.Coca AF. Specific Diagnosis and Treatment of Allergic Diseases of the Skin. JAMA. 1934;103(17):1275–1277. [Google Scholar]
- 4.Dold S, Wjst M, Mutius Ev, Reitmeir P, Stiepel E. Genetic risk for asthma, allergic rhinitis, and atopic dermatitis. Archives of Disease in Childhood. 1992;67:1018–1022. doi: 10.1136/adc.67.8.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sneddon IB. The management of infantile eczema. Vol. 226. Medical Press; 1951. pp. 329–333. [PubMed] [Google Scholar]
- 6.Schaffer N. Atopic dermatitis in the older child. The Journal of Asthma Research. 1966;3:189–191. doi: 10.3109/02770906609106918. [DOI] [PubMed] [Google Scholar]
- 7.Edfors-Lubs M-L. Allergy in 7000 twin pairs. Acta Allergology. 1971;26:249–285. doi: 10.1111/j.1398-9995.1971.tb01300.x. [DOI] [PubMed] [Google Scholar]
- 8.Schultz Larsen FV, Holm NV. Atopic dermatitis in a population based twin series. Concordance rates and heritability estimation. Acta Derm Venereol Suppl (Stockh) 1985;114:159. doi: 10.2340/00015555114159. [DOI] [PubMed] [Google Scholar]
- 9.Larsen FS, Holm NV, Henningsen K. Atopic dermatitis. A genetic-epidemiologic study in a population-based twin sample. J Am Acad Dermatol. 1986;15:487–494. [PubMed] [Google Scholar]
- 10.Strachan DP, Wong HJ, Spector TD. Concordance and interrelationship of atopic diseases and markers of allergic sensitization among adult female twins. J Allergy Clin Immunol. 2001;108:901–907. doi: 10.1067/mai.2001.119408. [DOI] [PubMed] [Google Scholar]
- 11.van Beijsterveldt CE, Boomsma DI. Genetics of parentally reported asthma, eczema and rhinitis in 5-yr-old twins. Eur Respir J. 2007;29:516–521. doi: 10.1183/09031936.00065706. [DOI] [PubMed] [Google Scholar]
- 12.Diepgen TL, Blettner M. Analysis of familial aggregation of atopic eczema and other atopic diseases by ODDS RATIO regression models. J Invest Dermatol. 1996;106:977–981. doi: 10.1111/1523-1747.ep12338475. [DOI] [PubMed] [Google Scholar]
- 13.Walley AJ, Chavanas S, Moffatt MF, Esnouf RM, Ubhi B, Lawrence R, et al. Gene polymorphism in Netherton and common atopic disease. Nature Genetics. 2001;29(2):175–178. doi: 10.1038/ng728. [DOI] [PubMed] [Google Scholar]
- 14.Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nature Genetics. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- 15.Lee YA, Wahn U, Kehrt R, Tarani L, Businco L, Gustafsson D, et al. A major susceptibility locus for atopic dermatitis maps to chromosome 3q21. Nature Genetics. 2000;26(4):470–473. doi: 10.1038/82625. [DOI] [PubMed] [Google Scholar]
- 16.Cookson WO, Ubhi B, Lawrence R, Abecasis GR, Walley AJ, Cox HE, et al. Genetic linkage of childhood atopic dermatitis to psoriasis susceptibility loci. Nature Genetics. 2001;27(4):372–373. doi: 10.1038/86867. [DOI] [PubMed] [Google Scholar]
- 17.Bradley M, Soderhall C, Luthman H, Wahlgren CF, Kockum I, Nordenskjold M. Susceptibility loci for atopic dermatitis on chromosomes 3, 13, 15, 17 and 18 in a Swedish population. Human Molecular Genetics. 2002;11(13):1539–1548. doi: 10.1093/hmg/11.13.1539. [DOI] [PubMed] [Google Scholar]
- 18.Haagerup A, Bjerke T, Schiotz PO, Dahl R, Binderup HG, Tan Q, et al. Atopic dermatitis -- a total genome-scan for susceptibility genes. Acta Derm Venereol. 2004;84:346–352. doi: 10.1080/00015550410034426. [DOI] [PubMed] [Google Scholar]
- 19.Guilloud-Bataille M, Bouzigon E, Annesi-Maesano I, Bousquet J, Charpin D, Gormand F, et al. Evidence for linkage of a new region (11p14) to eczema and allergic diseases. Hum Genet. 2008;122:605–614. doi: 10.1007/s00439-007-0439-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Enomoto H, Noguchi E, Iijima S, Takahashi T, Hayakawa K, Ito M, et al. Single nucleotide polymorphism-based genome-wide linkage analysis in Japanese atopic dermatitis families. BMC Dermatol. 2007;7:5. doi: 10.1186/1471-5945-7-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Christensen U, Moller-Larsen S, Nyegaard M, Haagerup A, Hedemand A, Brasch-Andersen C, et al. Linkage of atopic dermatitis to chromosomes 4q22, 3p24 and 3q21. Hum Genet. 2009 doi: 10.1007/s00439-009-0692-z. [DOI] [PubMed] [Google Scholar]
- 22.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 23.Ober C, Hoffjan S. Asthma genetics 2006: the long and winding road to gene discovery. Genes Immun. 2006 doi: 10.1038/sj.gene.6364284. [DOI] [PubMed] [Google Scholar]
- 24.Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 25.Neale BM, Sham PC. The future of association studies: gene-based analysis and replication. Am J Hum Genet. 2004;75:353–362. doi: 10.1086/423901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kabesch M. The art of replication. Thorax. 2009;64:370–371. doi: 10.1136/thx.2008.105197. [DOI] [PubMed] [Google Scholar]
- 27.International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morris AP. A flexible Bayesian framework for modeling haplotype association with disease, allowing for dominance effects of the underlying causative variants. Am J Hum Genet. 2006;79:679–694. doi: 10.1086/508264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rogers AJ, Raby BA, Lasky-Su JA, Murphy A, Lazarus R, Klanderman BJ, et al. Assessing the reproducibility of asthma candidate gene associations, using genome-wide data. Am J Respir Crit Care Med. 2009;179:1084–1090. doi: 10.1164/rccm.200812-1860OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldstein DB. Common genetic variation and human traits. N Engl J Med. 2009;360:1696–1698. doi: 10.1056/NEJMp0806284. [DOI] [PubMed] [Google Scholar]
- 31.Bodmer W, Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet. 2008;40:695–701. doi: 10.1038/ng.f.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gayan J, Gonzalez-Perez A, Bermudo F, Saez ME, Royo JL, Quintas A, et al. A method for detecting epistasis in genome-wide studies using case-control multi-locus association analysis. BMC Genomics. 2008;9:360. doi: 10.1186/1471-2164-9-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leung DY, Bieber T. Atopic dermatitis. Lancet. 2003;361:151–160. doi: 10.1016/S0140-6736(03)12193-9. [DOI] [PubMed] [Google Scholar]
- 34.Bieber T. Atopic dermatitis. N Engl J Med. 2008;358:1483–1494. doi: 10.1056/NEJMra074081. [DOI] [PubMed] [Google Scholar]
- 35.Beck LA, Boguniewicz M, Hata T, Schneider LC, Hanifin J, Gallo R, et al. Phenotype of subjects with atopic dermatitis with a history of eczema herpeticum. Journal of Allergy and Clinical Immunology. 2009;124:260–269. doi: 10.1016/j.jaci.2009.05.020. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hashimoto C, Hudson KL, Anderson KV. The Toll gene of Drosophila, required for dorsal-ventral embryonic polarity, appears to encode a transmembrane protein. Cell. 1988;52:269–279. doi: 10.1016/0092-8674(88)90516-8. [DOI] [PubMed] [Google Scholar]
- 37.Chamaillard M, Hashimoto M, Horie Y, Masumoto J, Qiu S, Saab L, et al. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat Immunol. 2003;4:702–707. doi: 10.1038/ni945. [DOI] [PubMed] [Google Scholar]
- 38.Girardin SE, Boneca IG, Carneiro LA, Antignac A, Jehanno M, Viala J, et al. Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science. 2003;300:1584–1587. doi: 10.1126/science.1084677. [DOI] [PubMed] [Google Scholar]
- 39.Bazil V, Horejsi V, Baudys M, Kristofova H, Strominger JL, Kostka W, et al. Biochemical characterization of a soluble form of the 53-kDa monocyte surface antigen. European Journal of Immunology. 1986;16(12):1583–1589. doi: 10.1002/eji.1830161218. [DOI] [PubMed] [Google Scholar]
- 40.Durieux JJ, Vita N, Popescu O, Guette F, Calzada-Wack J, Munker R, et al. The two soluble forms of the lipopolysaccharide receptor, CD14: characterization and release by normal human monocytes. European Journal of Immunology. 1994;24(9):2006–2012. doi: 10.1002/eji.1830240911. [DOI] [PubMed] [Google Scholar]
- 41.Labeta MO, Durieux JJ, Fernandez N, Herrmann R, Ferrara P. Release from a human monocyte-like cell line of two different soluble forms of the lipopolysaccharide receptor, CD14. European Journal of Immunology. 1993;23:2144. doi: 10.1002/eji.1830230915. [DOI] [PubMed] [Google Scholar]
- 42.Schroder JM, Harder J. Antimicrobial skin peptides and proteins. Cell Mol Life Sci. 2006;63:469–486. doi: 10.1007/s00018-005-5364-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raulet DH. Natural killer cells. In: Paul WE, editor. Fundamental Immunology. New York: Lipincott Williams-Wilkins; 2003. pp. 365–391. [Google Scholar]
- 44.McGirt LY, Beck LA. Innate immune defects in atopic dermatitis. J Allergy Clin Immunol. 2006;118:202–208. doi: 10.1016/j.jaci.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 45.De Benedetto A, Agnihothri R, McGirt LY, Bankova LG, Beck LA. Atopic dermatitis: a disease caused by innate immune defects? J Invest Dermatol. 2009;129:14–30. doi: 10.1038/jid.2008.259. [DOI] [PubMed] [Google Scholar]
- 46.Nomura I, Goleva E, Howell MD, Hamid QA, Ong PY, Hall CF, et al. Cytokine milieu of atopic dermatitis, as compared to psoriasis, skin prevents induction of innate immune response genes. J Immunol. 2003;171:3262–3269. doi: 10.4049/jimmunol.171.6.3262. [DOI] [PubMed] [Google Scholar]
- 47.Howell MD, Gallo RL, Boguniewicz M, Jones JF, Wong C, Streib JE, et al. Cytokine milieu of atopic dermatitis skin subverts the innate immune response to vaccinia virus. Immunity. 2006;24:341–348. doi: 10.1016/j.immuni.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 48.Kisich KO, Carspecken CW, Fieve S, Boguniewicz M, Leung DY. Defective killing of Staphylococcus aureus in atopic dermatitis is associated with reduced mobilization of human beta-defensin-3. J Allergy Clin Immunol. 2008;122:62–68. doi: 10.1016/j.jaci.2008.04.022. [DOI] [PubMed] [Google Scholar]
- 49.Leung DY. Infection in atopic dermatitis. Curr Opin Pediatr. 2003;15:399–404. doi: 10.1097/00008480-200308000-00008. [DOI] [PubMed] [Google Scholar]
- 50.Hauk PJ, Hamid QA, Chrousos GP, Leung DY. Induction of corticosteroid insensitivity in human PBMCs by microbial superantigens. J Allergy Clin Immunol. 2000;105:782–787. doi: 10.1067/mai.2000.105807. [DOI] [PubMed] [Google Scholar]
- 51.Gao L, Grant AV, Rafaels N, Stockton-Porter M, Watkins T, Gao P, et al. Polymorphisms in the myosin light chain kinase gene that confer risk of severe sepsis are associated with a lower risk of asthma. J Allergy Clin Immunol. 2007;119:1111–1118. doi: 10.1016/j.jaci.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 52.Levy H, Raby BA, Lake S, Tantisira KG, Kwiatkowski D, Lazarus R, et al. Association of defensin beta-1 gene polymorphisms with asthma. J Allergy Clin Immunol. 2005;115:252–258. doi: 10.1016/j.jaci.2004.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Matsushita I, Hasegawa K, Nakata K, Yasuda K, Tokunaga K, Keicho N. Genetic variants of human beta-defensin-1 and chronic obstructive pulmonary disease. Biochemical and Biophysical Research Communications. 2002;291:17–22. doi: 10.1006/bbrc.2002.6395. [DOI] [PubMed] [Google Scholar]
- 54.Segat L, Milanese M, Boniotto M, Crovella S, Bernardon M, Costantini M, et al. DEFB-1 genetic polymorphism screening in HIV-1 positive pregnant women and their children. J Matern Fetal Neonatal Med. 2006;19:13–16. doi: 10.1080/14767050500381123. [DOI] [PubMed] [Google Scholar]
- 55.Chen QX, Lv C, Huang LX, Cheng BL, Xie GH, Wu SJ, et al. Genomic variations within DEFB1 are associated with the susceptibility to and the fatal outcome of severe sepsis in Chinese Han population. Genes Immun. 2007;8:439–443. doi: 10.1038/sj.gene.6364401. [DOI] [PubMed] [Google Scholar]
- 56.Ferraris A, Torres B, Knafelz D, Barabino A, Lionetti P, de Angelis GL, et al. Relationship between CARD15, SLC22A4/5, and DLG5 polymorphisms and early-onset inflammatory bowel diseases: an Italian multicentric study. Inflamm Bowel Dis. 2006;12:355–361. doi: 10.1097/01.MIB.0000217338.23065.58. [DOI] [PubMed] [Google Scholar]
- 57.Gazouli M, Koundourakis A, Ikonomopoulos J, Gialafos EJ, Rapti A, Gorgoulis VG, et al. CARD15/NOD2, CD14, and toll-like receptor 4 gene polymorphisms in Greek patients with sarcoidosis. Sarcoidosis Vasc Diffuse Lung Dis. 2006;23:23–29. [PubMed] [Google Scholar]
- 58.Cork MJ, Robinson DA, Vasilopoulos Y, Ferguson A, Moustafa M, MacGowan A, et al. New perspectives on epidermal barrier dysfunction in atopic dermatitis: gene-environment interactions. J Allergy Clin Immunol. 2006;118:3–21. doi: 10.1016/j.jaci.2006.04.042. quiz 2–3. [DOI] [PubMed] [Google Scholar]
- 59.Callard RE, Harper JI. The skin barrier, atopic dermatitis and allergy: a role for Langerhans cells? Trends Immunol. 2007;28:294–298. doi: 10.1016/j.it.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 60.Holgate ST. Epithelium dysfunction in asthma. J Allergy Clin Immunol. 2007;120:1233–1244. doi: 10.1016/j.jaci.2007.10.025. quiz 45–46. [DOI] [PubMed] [Google Scholar]
- 61.Holgate ST. The epithelium takes centre stage in asthma and atopic dermatitis. Trends Immunol. 2007;28:248–251. doi: 10.1016/j.it.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 62.Elias PM, Hatano Y, Williams ML. Basis for the barrier abnormality in atopic dermatitis: outside-inside-outside pathogenic mechanisms. J Allergy Clin Immunol. 2008;121:1337–1343. doi: 10.1016/j.jaci.2008.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hammad H, Lambrecht BN. Dendritic cells and epithelial cells: linking innate and adaptive immunity in asthma. Nat Rev Immunol. 2008;8:193–204. doi: 10.1038/nri2275. [DOI] [PubMed] [Google Scholar]
- 64.Higaki S, Morohashi M, Yamagishi T, Hasegawa Y. Comparative study of staphylococci from the skin of atopic dermatitis patients and from healthy subjects. Int J Dermatol. 1999;38:265–269. doi: 10.1046/j.1365-4362.1999.00686.x. [DOI] [PubMed] [Google Scholar]
- 65.Travers JB, Norris DA, Leung DY. The keratinocyte as a target for staphylococcal bacterial toxins. J Investig Dermatol Symp Proc. 2001;6:225–230. doi: 10.1046/j.0022-202x.2001.00045.x. [DOI] [PubMed] [Google Scholar]
- 66.Ricci G, Patrizi A, Neri I, Bendandi B, Masi M. Frequency and clinical role of Staphylococcus aureus overinfection in atopic dermatitis in children. Pediatr Dermatol. 2003;20:389–392. doi: 10.1046/j.1525-1470.2003.20503.x. [DOI] [PubMed] [Google Scholar]
- 67.Sugimoto K, Ishikawa N, Terano T, Kitukawa Y, Kubosawa H, Ito S, et al. The importance of bacterial superantigens produced by Staphylococcus aureus in the treatment of atopic dermatitis using povidone-iodine. Dermatology. 2006;212 Suppl 1:26–34. doi: 10.1159/000089196. [DOI] [PubMed] [Google Scholar]
- 68.Warner JA, McGirt LY, Beck LA. Biomarkers of Th2 polarity are predictive of staphylococcal colonization in subjects with atopic dermatitis. Br J Dermatol. 2009;160:183–185. doi: 10.1111/j.1365-2133.2008.08905.x. [DOI] [PubMed] [Google Scholar]
- 69.Niessen CM. Tight junctions/adherens junctions: basic structure and function. J Invest Dermatol. 2007;127:2525–2532. doi: 10.1038/sj.jid.5700865. [DOI] [PubMed] [Google Scholar]
- 70.Aijaz S, D'Atri F, Citi S, Balda MS, Matter K. Binding of GEF-H1 to the tight junction-associated adaptor cingulin results in inhibition of Rho signaling and G1/S phase transition. Dev Cell. 2005;8:777–786. doi: 10.1016/j.devcel.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 71.Sawada N, Murata M, Kikuchi K, Osanai M, Tobioka H, Kojima T, et al. Tight junctions and human diseases. Med Electron Microsc. 2003;36:147–156. doi: 10.1007/s00795-003-0219-y. [DOI] [PubMed] [Google Scholar]
- 72.Hoffjan S, Stemmler S. On the role of the epidermal differentiation complex in ichthyosis vulgaris, atopic dermatitis and psoriasis. Br J Dermatol. 2007;157:441–449. doi: 10.1111/j.1365-2133.2007.07999.x. [DOI] [PubMed] [Google Scholar]
- 73.O'Regan GM, Sandilands A, McLean WH, Irvine AD. Filaggrin in atopic dermatitis. J Allergy Clin Immunol. 2008;122:689–693. doi: 10.1016/j.jaci.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 74.McKinley-Grant LJ, Idler WW, Bernstein IA, Parry DA, Cannizzaro L, Croce CM, et al. Characterization of a cDNA clone encoding human filaggrin and localization of the gene to chromosome region 1q21. Proc Natl Acad Sci U S A. 1989;86:4848–4852. doi: 10.1073/pnas.86.13.4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gan SQ, McBride OW, Idler WW, Markova N, Steinert PM. Organization, structure, and polymorphisms of the human profilaggrin gene. Biochemistry. 1990;29:9432–9440. doi: 10.1021/bi00492a018. [DOI] [PubMed] [Google Scholar]
- 76.Smith FJ, Irvine AD, Terron-Kwiatkowski A, Sandilands A, Campbell LE, Zhao Y, et al. Loss-of-function mutations in the gene encoding filaggrin cause ichthyosis vulgaris. Nat Genet. 2006;38:337–342. doi: 10.1038/ng1743. [DOI] [PubMed] [Google Scholar]
- 77.Palmer CN, Irvine AD, Terron-Kwiatkowski A, Zhao Y, Liao H, Lee SP, et al. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat Genet. 2006;38:441–446. doi: 10.1038/ng1767. [DOI] [PubMed] [Google Scholar]
- 78.Sandilands A, Terron-Kwiatkowski A, Hull PR, O'Regan GM, Clayton TH, Watson RM, et al. Comprehensive analysis of the gene encoding filaggrin uncovers prevalent and rare mutations in ichthyosis vulgaris and atopic eczema. Nat Genet. 2007;39:650–654. doi: 10.1038/ng2020. [DOI] [PubMed] [Google Scholar]
- 79.Rodriguez E, Baurecht H, Herberich E, Wagenpfeil S, Brown SJ, Cordell HJ, et al. Meta-analysis of filaggrin polymorphisms in eczema and asthma: robust risk factors in atopic disease. J Allergy Clin Immunol. 2009;123:1361–1370. doi: 10.1016/j.jaci.2009.03.036. e7. [DOI] [PubMed] [Google Scholar]
- 80.Palmer CN, Ismail T, Lee SP, Terron-Kwiatkowski A, Zhao Y, Liao H, et al. Filaggrin null mutations are associated with increased asthma severity in children and young adults. J Allergy Clin Immunol. 2007;120:64–68. doi: 10.1016/j.jaci.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 81.Novak N, Baurecht H, Schafer T, Rodriguez E, Wagenpfeil S, Klopp N, et al. Loss-of-function mutations in the filaggrin gene and allergic contact sensitization to nickel. J Invest Dermatol. 2008;128:1430–1435. doi: 10.1038/sj.jid.5701190. [DOI] [PubMed] [Google Scholar]
- 82.Ekelund E, Lieden A, Link J, Lee SP, D'Amato M, Palmer CN, et al. Loss-of-function variants of the filaggrin gene are associated with atopic eczema and associated phenotypes in Swedish families. Acta Derm Venereol. 2008;88:15–19. doi: 10.2340/00015555-0383. [DOI] [PubMed] [Google Scholar]
- 83.Basu K, Palmer CN, Lipworth BJ, Irwin McLean WH, Terron-Kwiatkowski A, Zhao Y, et al. Filaggrin null mutations are associated with increased asthma exacerbations in children and young adults. Allergy. 2008;63:1211–1217. doi: 10.1111/j.1398-9995.2008.01660.x. [DOI] [PubMed] [Google Scholar]
- 84.Henderson J, Northstone K, Lee SP, Liao H, Zhao Y, Pembrey M, et al. The burden of disease associated with filaggrin mutations: a population-based, longitudinal birth cohort study. J Allergy Clin Immunol. 2008;121:872–877. doi: 10.1016/j.jaci.2008.01.026. e9. [DOI] [PubMed] [Google Scholar]
- 85.Weidinger S, O'Sullivan M, Illig T, Baurecht H, Depner M, Rodriguez E, et al. Filaggrin mutations, atopic eczema, hay fever, and asthma in children. J Allergy Clin Immunol. 2008;121:1203–1209. doi: 10.1016/j.jaci.2008.02.014. e1. [DOI] [PubMed] [Google Scholar]
- 86.de Jongh CM, Khrenova L, Verberk MM, Calkoen F, van Dijk FJ, Voss H, et al. Loss-of-function polymorphisms in the filaggrin gene are associated with an increased susceptibility to chronic irritant contact dermatitis: a case-control study. Br J Dermatol. 2008 doi: 10.1111/j.1365-2133.2008.08730.x. [DOI] [PubMed] [Google Scholar]
- 87.Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- 88.Esparza-Gordillo J, Weidinger S, Folster-Holst R, Bauerfeind A, Ruschendorf F, Patone G, et al. A common variant on chromosome 11q13 is associated with atopic dermatitis. Nat Genet. 2009;41:596–601. doi: 10.1038/ng.347. [DOI] [PubMed] [Google Scholar]
- 89.Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sugiura H, Ebise H, Tazawa T, Tanaka K, Sugiura Y, Uehara M, et al. Large-scale DNA microarray analysis of atopic skin lesions shows overexpression of an epidermal differentiation gene cluster in the alternative pathway and lack of protective gene expression in the cornified envelope. Br J Dermatol. 2005;152:146–149. doi: 10.1111/j.1365-2133.2005.06352.x. [DOI] [PubMed] [Google Scholar]
- 91.Lu ZR, Park D, Lee KA, Ryu JW, Bhak J, Shi L, et al. Profiling the dysregulated genes of keratinocytes in atopic dermatitis patients: cDNA microarray and interactomic analyses. J Dermatol Sci. 2009;54:126–129. doi: 10.1016/j.jdermsci.2008.12.006. [DOI] [PubMed] [Google Scholar]
- 92.Toulza E, Mattiuzzo NR, Galliano MF, Jonca N, Dossat C, Jacob D, et al. Large-scale identification of human genes implicated in epidermal barrier function. Genome Biol. 2007;8:R107. doi: 10.1186/gb-2007-8-6-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 94.Morgan JK. Sensitization and eczema. Occupational Medicine (London) 1953;3:274–282. [Google Scholar]
- 95.Gereda JE, Leung DY, Thatayatikom A, Streib JE, Price MR, Klinnert MD, et al. Relation between house-dust endotoxin exposure, type 1 T-cell development, and allergen sensitisation in infants at high risk of asthma. Lancet. 2000;355(9216):1680–1683. doi: 10.1016/s0140-6736(00)02239-x. [DOI] [PubMed] [Google Scholar]
- 96.Braun-Fahrlander C, Riedler J, Herz U, Eder W, Waser M, Grize L, et al. Environmental exposure to endotoxin and its relation to asthma in school-age children. New England Journal of Medicine. 2002;347(12):869–877. doi: 10.1056/NEJMoa020057. [DOI] [PubMed] [Google Scholar]
- 97.Strachan DP. Hay fever, hygiene, and household size. British Medical Journal. 1989;299(6710):1259–1260. doi: 10.1136/bmj.299.6710.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sherriff A, Golding J. Hygiene levels in a contemporary population cohort are associated with wheezing and atopic eczema in preschool infants. Arch Dis Child. 2002;87:26–29. doi: 10.1136/adc.87.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bisgaard H, Simpson A, Palmer CN, Bonnelykke K, McLean I, Mukhopadhyay S, et al. Gene-environment interaction in the onset of eczema in infancy: filaggrin loss-of-function mutations enhanced by neonatal cat exposure. PLoS Medicine. 2008;5:e131. doi: 10.1371/journal.pmed.0050131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.HR A. The epidemiological and allergic features of asthma in the New Guinea Highlands. Clinical Allergy. 1974;4(2):171–183. doi: 10.1111/j.1365-2222.1974.tb01374.x. 171-83 CAJ. [DOI] [PubMed] [Google Scholar]
- 101.Ioannidis JP, Ntzani EE, Trikalinos TA, Contopoulos-Ioannidis DG. Replication validity of genetic association studies. Nat Genet. 2001;29:306–309. doi: 10.1038/ng749. [DOI] [PubMed] [Google Scholar]
- 102.Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genetic Medicine. 2002;4(2):45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- 103.Colhoun HM, McKeigue PM, Davey Smith G. Problems of reporting genetic associations with complex outcomes. Lancet. 2003;361:865–872. doi: 10.1016/s0140-6736(03)12715-8. [DOI] [PubMed] [Google Scholar]
- 104.Janumpally SR, Feldman SR, Gupta AK, Fleischer ABJ. In the United States, blacks and Asian/Pacific Islanders are more likely than whites to seek medical care for atopic dermatitis. Archives of Dermatology. 2002;138(5) doi: 10.1001/archderm.138.5.634. [DOI] [PubMed] [Google Scholar]
- 105.Nomura T, Sandilands A, Akiyama M, Liao H, Evans AT, Sakai K, et al. Unique mutations in the filaggrin gene in Japanese patients with ichthyosis vulgaris and atopic dermatitis. J Allergy Clin Immunol. 2007;119:434–440. doi: 10.1016/j.jaci.2006.12.646. [DOI] [PubMed] [Google Scholar]
- 106.Giardina E, Paolillo N, Sinibaldi C, Novelli G. R501X and 2282del4 filaggrin mutations do not confer susceptibility to psoriasis and atopic dermatitis in Italian patients. Dermatology. 2008;216:83–84. doi: 10.1159/000109365. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.