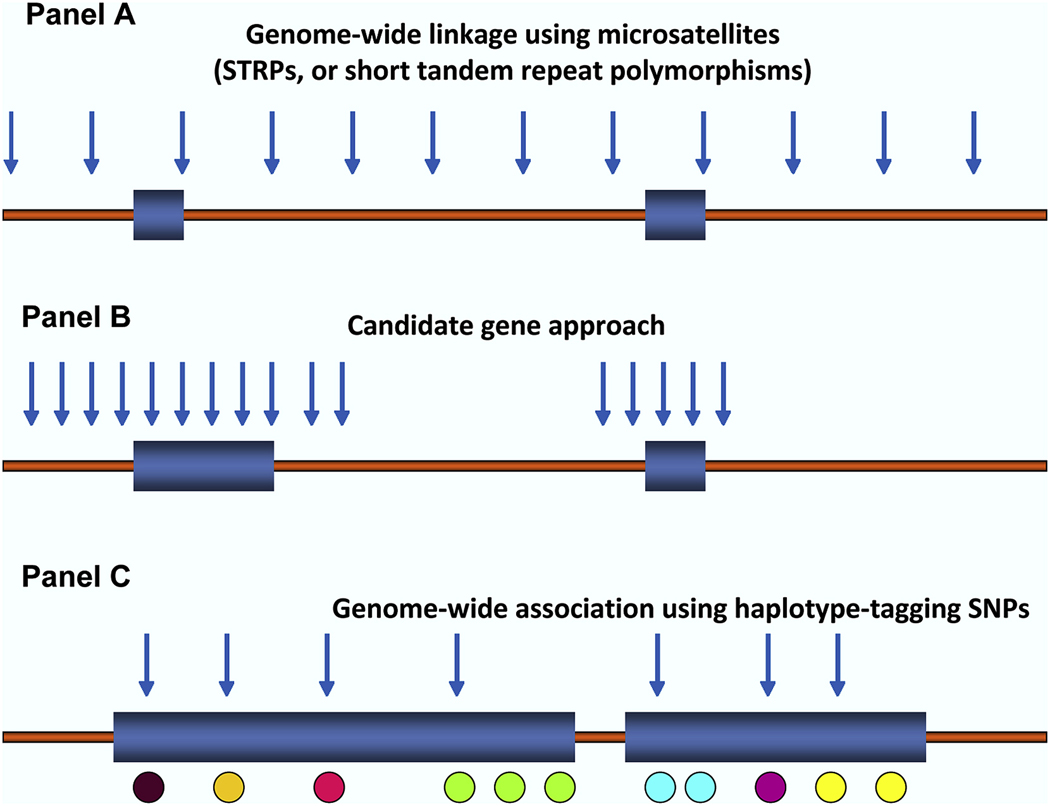
Figure 1. Methods for detecting disease susceptibility genes.
Panel A, genome-wide linkage method, relying on polymorphic microsatellites, referred to as ‘short tandem repeat polymorphisms’, or STRPs, evenly spaced across the chromosomes, typically including ~350 STRPs. Panel B, the candidate gene approach, whereby genetic markers, usually easily typed substitutions (i.e., single nucleotide polymorphisms, or SNPs) or structural variants (i.e., insertions/deletions), are selected within and flanking a candidate gene of interest. Panel C, genome-wide association studies, or GWAS, using hundreds of thousands and up to 1M SNPs to fully cover an individual’s genome, is a hypothesis-free, systematic search across the genome to identify novel associations with common diseases using a set of haplotype tagging SNPs (htSNPs), indicated as blue arrows. htSNPs represent a relatively small subset of SNPs of the potential 2.5 million SNPs in the public database (i.e., needed to uniquely identify a complete haplotype. The premise behind htSNPs is the observation that, when SNPs are in linkage disequilibrium (LD) with each other and form haplotypes, there is redundant information contained within the haplotype, because several of the SNPs will always occur together. Thus, a marker at one locus can reasonably predict a marker(s) that will occur at the linked locus nearby.