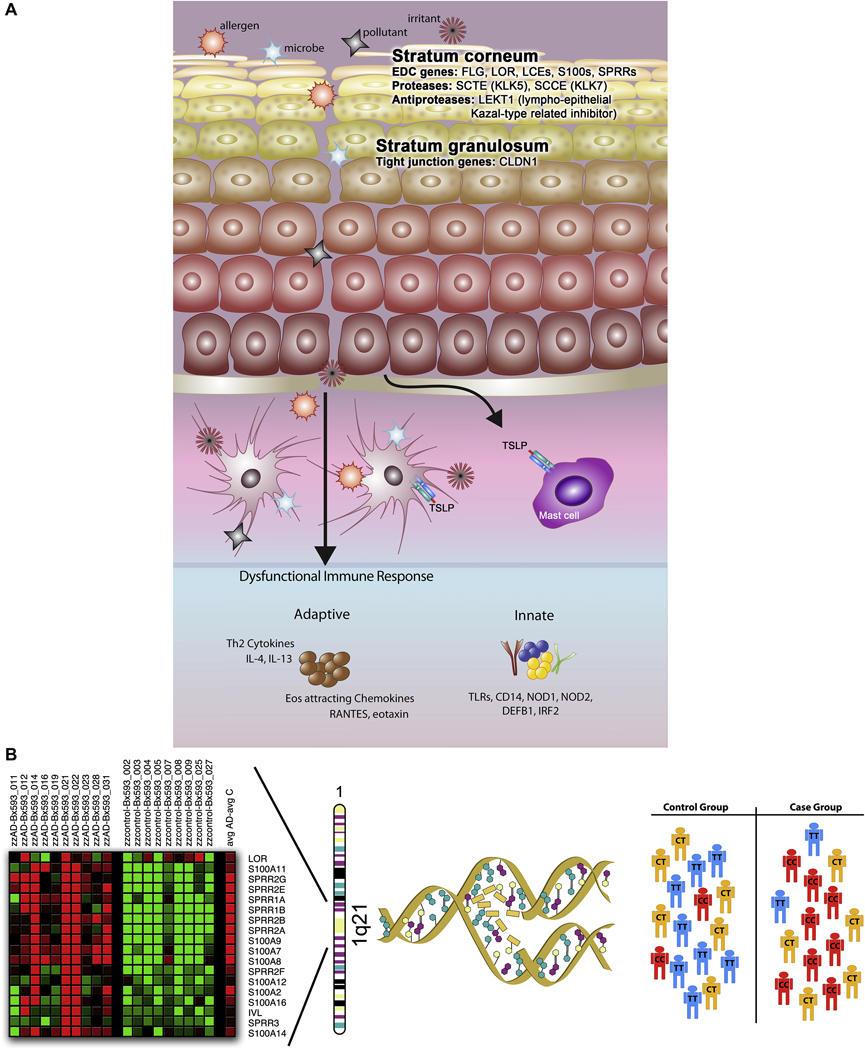
Figure 5. Genetic epidemiological approaches toward identification of genes conferring AD susceptibility.
In this summary of approaches used to date to identify candidate genes for AD, the first consideration is the complex pathology of AD (panel A), which includes defects and damage at the epithelial barrier, and penetration of allergens, microbes, pollutants and irritants into the epidermis and dermis, ultimately interacting with antigen presentation cells (i.e., Langerhans cells and dermal dendritic cells). The ‘brick wall-like’ structure of the stratum corneum is compromised, possibly by defects in genes residing in the epidermal differentiation complex (EDC), including FLG, LOR, LCEs, S100s, SPRRs, SCTE, and SCCE. Additional candidates include tight junctions (TJ), proteins that constitute the “gate” to the passage of water, ions and solutes through the paracellular pathway in the stratum granulosum (i.e., CLDN1). A defective innate immune response (involving, for example, the TLRs, CD14, NOD1, NOD2, DEFB1, and IRF2) may contribute to a heightened IgE-mediated, systemic Th2 response. Combining a candidate gene approach with robust, genomewide gene expression profiling has the potential of both elucidating novel candidates and validating suspected candidates, as illustrated in panel B, in which genes in the EDC on chromosome 1q21 are significantly differentially expressed in a human skin biopsies taken from AD patients compared to healthy nonatopic controls (courtesy L.A. Beck). Candidate genes are selected, and substitutions (i.e., single nucleotide polymorphisms, or SNPs) and simple structural variants (i.e., insertions/deletions, repeats) are genotyped in large populations selected for AD and tests for association are performed.