Figure.
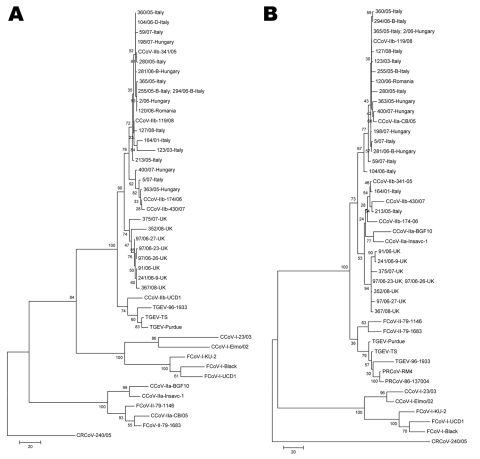
Phylogenetic analysis of canine coronavirus (CCoV) type IIb. Maximum parsimony trees based on partial 5´ (A) and 3´ (B) ends of the spike protein gene of group-1a coronaviruses (CoVs). For phylogenetic tree construction, the following reference strains were used (GenBank accession numbers are in parentheses): porcine transmissible gastroenteritis virus (TGEV) Purdue (NC_002306), TS (DQ201447), 96-1933 (AF104420), porcine respiratory coronavirus (PRCoV, only 3´ end) RM-4 (Z24675), 86-137004 (X60089), CCoV-IIa CB/05 (DQ112226), Insavc-1 (D13096), BGF10 (AY342160), CCoV-IIb 341/05 (EU856361), 174/06 (EU856362), 430/07 (EU924790), 118/08 (EU924791), UCD-1 (AF116248, only 5´ end), CCoV-I Elmo/02 (AY307020), 23/03 (AY307021), feline coronavirus (FCoV) type I Black (EU186072), KU-1 (D32044), UCD-1 (AB088222), FCoV-II 79-1146 (NC_007025), 79-1183 (X80799). The tree is rooted on the group-2 CoV canine respiratory coronavirus (CRCoV) 240/05 (EU999954). Statistical support was provided by bootstrapping >1,000 replicates. Scale bars indicate estimated numbers of nucleotide substitutions per site.