Abstract
The epithelial transmembrane glycoprotein E-cadherin (CDH1) is necessary for intercellular adhesion, cell signaling, and maintenance of cellular differentiation; reduced expression contributes to cell proliferation, invasion, and cancer progression. Functional or potentially functional single nucleotide polymorphisms (SNPs) in E-cadherin have been previously identified and evaluated in relation to cancer risk; however, studies on breast cancer have been sparse. Forty-six SNPs were genotyped to capture genetic variation of the CDH1 gene among 2,290 Phase 1 and 1,944 Phase 2 participants of the Shanghai Breast Cancer Study (SBCS), a large, population-based, case–control study. No overall associations between E-cadherin SNPs and breast cancer risk were observed. When stratified by menopausal status, associations that were consistent between Phases 1 and 2 and significant when data from both phases were combined were observed for several SNPs. Although none of these associations retained statistical significance after correcting for the total number of polymorphisms evaluated, this study suggests that genetic variation in CDH1 may be associated with breast cancer risk, and that this relationship may vary by menopausal status.
Keywords: E-cadherin, Polymorphisms, Breast cancer risk
Introduction
The transmembrane glycoprotein E-cadherin (CDH1) is necessary for normal epithelial cells intercellular adhesion, cell polarity, cell signaling, and maintenance of cellular differentiation and tissue morphology [1–3]. Diminished E-cadherin expression promotes malignant transformation, tumor invasion, and metastasis [1–3]. A promoter polymorphism (−160 C/A, rs16260) that results in reduced E-cadherin expression for the minor allele (A) [4–6] has been extensively studied and is suggested to be associated with increased susceptibility to lung, prostate, and gastric cancers in meta-analyses [7, 8]. In regards to breast cancer, only two case–control studies have evaluated this single nucleotide polymorphism (SNP); one found no association [5], while the other found a significantly increased risk among A allele carriers [9]. Additional promoter polymorphisms that influence E-cadherin expression have also been reported (−347 G/GA, −288 T/−, −285 C/A, −54 G/C) [10, 11], as have other potentially functional SNPs (163 + 37235 G/A in intron 2, and 3′ UTR + 54 C/T) [12, 13], but none have been evaluated in relation to breast cancer risk. This study was, therefore, undertaken to comprehensively assess individual genetic variation across E-cadherin, and evaluate associations with susceptibility to breast cancer among participants of the Shanghai Breast Cancer Study (SBCS).
Methods
Study subjects were participants of the SBCS, a large, two-phase, population-based, case–control study of women in urban Shanghai which has been previously described in detail [14–16]. Briefly, breast cancer cases were identified via a rapid case-ascertainment system in Phase 1, and the Shanghai Cancer Registry in Phases 1 and 2; diagnoses were confirmed by two senior pathologists. Controls were randomly selected using the Shanghai Resident Registry. Phase 1 recruitment occurred between August 1996 and March 1998, and included women aged 25–65. Phase 2 recruitment occurred from April 2002 to February 2005 and was expanded to include women aged 20–70. In-person interviews were completed for 1,459 (91.1%) cases and 1,556 (90.3%) controls from Phase 1, and 1,989 cases (83.7%) and 1,989 controls (70.4%) from Phase 2. Blood samples were donated by 1,193 cases (81.8%) and 1,310 controls (84.2%) from Phase 1 and blood or buccal cell samples were donated by 1,932 (97.1%) cases and 1,857 (93.4%) controls from Phase 2. Approval was granted from relevant review boards in both China and the United States.
Haplotype tagging SNPs (htSNPs) were selected using Han Chinese data presented in the HapMap Project [17] using the Tagger program [18] to capture SNPs with a minimum minor allele frequency (MAF) of 0.05 in E-cadherin (±5 kb) with an r2 of 0.90 or greater. Twenty-eight E-cadherin SNPs were selected; twenty-four were successfully designed and genotyped in 2006 for 1,062 cases and 1,069 controls from Phase 1, using a Targeted Genotyping System (Affymetrix, Santa Clara, CA) as previously described [15]. In order to increase the density of genetic markers in this study, data from our recently completed Affymetrix Genome-Wide Human SNP Array 6.0 (Affymetrix) was included for an additional 22 E-cadherin polymorphisms (±10 kb) that were genotyped among 1,104 cases and 1,109 controls from Phase 1, and 969 cases and 975 controls from Phase 2.
Hardy–Weinberg equilibrium (HWE) was tested by comparing the observed and expected genotype frequencies of the controls (χ2-test). Odds ratios (ORs) and corresponding 95% confidence intervals (CIs) were determined by logistic regression analyses using models that included adjustment for age, education, and study phase if appropriate. Additive, dominant, recessive, and allelic associations were considered. Linkage disequilibrium (LD) was assessed by Haploview [19]. Haplotype analysis was conducted with Hapstat [20]. Initial statistical significance was determined with a threshold P value of 0.05; however, to address the issue of multiple comparisons, the Bonferroni correction was then employed. All statistical tests were two-tailed.
Results
A total of 4,234 women were included in this study: 2,290 Phase 1 participants and 1,944 Phase 2 participants (Table 1). Women in both study phases were generally comparable. As expected, breast cancer cases were found to differ from controls in regards to known breast cancer risk factors; cases were more likely to have earlier age at menarche, older age at first live birth, a history of breast fibroadenomas, a history of breast cancer among a first degree relative, a higher body mass index (BMI) and/or waist-to-hip ratio (WHR), and less likely to participate in regular physical activity than controls.
Table 1.
Shanghai Breast Cancer Study participants genotyped for E-cadherin, the Shanghai Breast Cancer Study
| Characteristics | Phase 1 (N = 2,290) |
Phase 2 (N = 1,944) |
||||
|---|---|---|---|---|---|---|
| Cases (N = 1,114) | Controls (N = 1,176) | P value | Cases (N = 969) | Controls (N = 975) | P value | |
| Demographic factors | ||||||
| Age (years) | 47.6 ± 8.0 | 47.6 ± 8.3 | 0.807 | 51.4 ± 8.3 | 51.4 ± 8.2 | 0.913 |
| Education (less than middle school) | 138 (12.4%) | 171 (14.5%) | 0.132 | 69 (7.1%) | 115 (11.8%) | < 0.001 |
| Reproductive risk factors | ||||||
| Age at menarche (years) | 14.5 ± 1.6 | 14.7 ± 1.7 | < 0.001 | 14.5 ± 1.7 | 14.7 ± 1.8 | 0.004 |
| Premenopausal | 742 (67.0%) | 704 (63.3%) | 0.071 | 528 (54.5%) | 516 (52.9%) | 0.489 |
| Age at menopause (years)a | 48.1 ± 4.7 | 47.4 ± 5.0 | 0.034 | 48.7 ± 4.4 | 48.0 ± 4.5 | 0.027 |
| Age at first live birth (years)b | 26.8 ± 4.1 | 26.3 ± 3.8 | 0.001 | 26.1 ± 3.7 | 25.7 ± 3.8 | 0.027 |
| Used oral contraceptives | 244 (22.0%) | 253 (21.6%) | 0.811 | 175 (18.1%) | 180 (18.5%) | 0.819 |
| Used estrogen replacement therapy | 28 (2.5%) | 30 (2.6%) | 0.963 | 37 (3.8%) | 21 (2.2%) | 0.031 |
| Additional risk factors | ||||||
| First degree relative with breast cancer | 37 (3.3%) | 30 (2.6%) | 0.272 | 55 (5.7%) | 33 (3.4%) | 0.015 |
| Ever had breast fibroadenomas | 105 (9.5%) | 58 (5.0%) | < 0.001 | 96 (10.0%) | 63 (6.5%) | 0.005 |
| Body mass index (kg/m2) | 23.6 ± 3.4 | 23.2 ± 3.4 | 0.013 | 24.0 ± 3.3 | 23.4 ± 3.3 | < 0.001 |
| Waist-to-hip ratio | 0.81 ± 0.06 | 0.80 ± 0.06 | 0.002 | 0.84 ± 0.05 | 0.82 ± 0.06 | < 0.001 |
| Regular physical activity | 213 (19.2%) | 300 (25.6%) | < 0.001 | 308 (31.8%) | 338 (34.7%) | 0.178 |
Continuous variables: mean values ± standard deviation, P value from t-tests; Categorical variables: numbers and percentages, P values from χ2 test
Among postmenopausal women
Among parous women
Bold values considered to be significant P ≤ 0.05
A total of 46 E-cadherin SNPs were included in this study: 24 htSNPs and 22 additional SNPs from the Affymetrix Genome-Wide Human SNP Array 6.0 (Affymetrix). Of these polymorphisms, none were found to deviate from HWE, but six were found to have minor allele frequencies (MAFs) of less than 5% (rs7194684, rs3931740, rs8049967, rs7190460, rs13330170, and rs2276328). Eleven SNPs were included by both genotyping methods included in this study for an average of 1,895 participants; concordance rates for these samples ranged from 99.6–100, and averaged 99.78. When two genotyping sources were available for one polymorphism, the source with the larger number of samples genotyped was used in our analyses. Information and estimates of effect for the 40 E-cadherin polymorphisms with MAF ≥ 5% are shown in Table 2. In analyses including all women, no SNPs were found to be significantly associated with breast cancer risk in additive models that included adjustment for age, education, and study phase (when appropriate). Further, no significant associations were identified under dominant or recessive models (data not shown). For simplification, estimates of effect on risk per minor allele are presented.
Table 2.
E-cadherin SNPs and breast cancer risk among all women, premenopausal women, and postmenopausal women, the Shanghai Breast Cancer Study
| SNP | Allelesa | Region | Genotypingb | MAFc | HWEd | All women |
Premenopausal womenf |
Postmenopausal womeng |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|
| B OR (95% CI)e | P valuee | B OR (95% CI)e | P valuee | B OR (95% CI)e | P value | ||||||
| rs9989407 | T/C | Promoter | Affy 6.0 | 25.8 | 0.560 | 1.1 (1.0–1.2) | 0.334 | 1.0 (0.9–1.1) | 0.673 | 1.2 (1.0–1.4) | 0.045 |
| rs9940250 | T/C | Promoter | Targeted | 27.9 | 0.905 | 1.0 (0.8–1.1) | 0.594 | 0.9 (0.8–1.1) | 0.427 | 1.0 (0.8–1.3) | 0.849 |
| rs7196495 | T/C | Intron 2 | Affy 6.0 | 25.9 | 0.630 | 1.0 (0.9–1.1) | 0.409 | 1.0 (0.9–1.1) | 0.635 | 1.2 (1.0–1.3) | 0.065 |
| rs7196661 | T/C | Intron 2 | Affy 6.0 | 25.9 | 0.630 | 1.0 (0.9–1.1) | 0.416 | 1.0 (0.9–1.1) | 0.637 | 1.2 (1.0–1.3) | 0.069 |
| rs11865026 | T/C | Intron 2 | Affy 6.0 | 24.2 | 0.976 | 1.0 (0.9–1.1) | 0.671 | 1.0 (0.9–1.2) | 0.627 | 0.9 (0.8–1.1) | 0.216 |
| rs7203337 | G/C | Intron 2 | Affy 6.0 | 49.8 | 0.630 | 1.0 (0.9–1.1) | 0.724 | 1.0 (0.9–1.1) | 0.999 | 1.0 (0.8–1.1) | 0.569 |
| rs1078621 | C/T | Intron 2 | Affy 6.0 | 49.2 | 0.155 | 1.0 (0.9–1.1) | 0.449 | 1.0 (0.9–1.1) | 0.923 | 0.9 (0.8–1.0) | 0.177 |
| rs11642413 | G/A | Intron 2 | Targeted | 48.4 | 0.803 | 1.0 (0.9–1.2) | 0.658 | 1.0 (0.9–1.2) | 0.697 | 1.0 (0.8–1.3) | 0.856 |
| rs9941051 | T/C | Intron 2 | Affy 6.0 | 25.7 | 0.423 | 1.0 (0.9–1.1) | 0.428 | 1.0 (0.9–1.1) | 0.712 | 1.1 (1.0–1.3) | 0.097 |
| rs8056538 | G/A | Intron 2 | Affy 6.0 | 24.1 | 0.804 | 1.0 (0.9–1.1) | 0.949 | 1.1 (0.9–1.2) | 0.417 | 0.9 (0.8–1.1) | 0.280 |
| rs12444784 | A/G | Intron 2 | Both | 8.2 | 0.069 | 0.9 (0.8–1.1) | 0.352 | 0.8 (0.7–1.0) | 0.058 | 1.1 (0.9–1.5) | 0.342 |
| rs12930371 | C/T | Intron 2 | Affy 6.0 | 24.1 | 0.548 | 1.0 (0.9–1.1) | 0.966 | 1.1 (0.9–1.2) | 0.308 | 0.9 (0.8–1.1) | 0.283 |
| rs2113200 | T/A | Intron 2 | Affy 6.0 | 24.5 | 0.864 | 1.0 (0.9–1.1) | 0.723 | 1.0 (0.9–1.2) | 0.554 | 0.9 (0.8–1.1) | 0.203 |
| rs9929498 | G/A | Intron 2 | Targeted | 26.4 | 0.729 | 1.0 (0.9–1.1) | 0.841 | 1.0 (0.8–1.1) | 0.686 | 1.0 (0.8–1.3) | 0.918 |
| rs2059254 | C/T | Intron 2 | Both | 16.6 | 0.469 | 1.0 (0.9–1.2) | 0.690 | 1.2 (1.0–1.4) | 0.029 | 0.8 (0.7–1.0) | 0.049 |
| rs9925923 | C/T | Intron 2 | Affy 6.0 | 17.1 | 0.578 | 1.0 (0.9–1.2) | 0.678 | 1.2 (1.0–1.4) | 0.024 | 0.8 (0.7–1.0) | 0.049 |
| rs12919719 | C/G | Intron 2 | Both | 17.6 | 0.792 | 1.0 (0.9–1.1) | 0.913 | 1.2 (1.0–1.4) | 0.057 | 0.8 (0.7–1.0) | 0.044 |
| rs4076177 | A/G | Intron 2 | Targeted | 20.8 | 0.668 | 1.1 (0.9–1.2) | 0.508 | 1.1 (0.9–1.3) | 0.307 | 1.0 (0.8–1.3) | 0.904 |
| rs12599393 | C/T | Intron 2 | Affy 6.0 | 17.1 | 0.638 | 1.0 (0.9–1.1) | 0.752 | 1.1 (1.0–1.3) | 0.129 | 0.8 (0.7–1.0) | 0.020 |
| rs1862748 | C/T | Intron 2 | Both | 18.1 | 0.707 | 1.0 (0.9–1.1) | 0.834 | 1.1 (1.0–1.3) | 0.100 | 0.8 (0.7–1.0) | 0.027 |
| rs10431923 | T/G | Intron 2 | Affy 6.0 | 45.2 | 0.311 | 1.0 (1.0–1.2) | 0.424 | 1.1 (1.0–1.2) | 0.222 | 1.0 (0.9–1.1) | 0.875 |
| rs10431924 | C/T | Intron 2 | Targeted | 44.7 | 0.735 | 1.0 (0.9–1.2) | 0.631 | 1.1 (0.9–1.2) | 0.355 | 1.0 (0.8–1.2) | 0.703 |
| rs4783573 | G/A | Intron 2 | Both | 21.9 | 0.370 | 1.0 (0.9–1.1) | 0.545 | 1.0 (0.9–1.2) | 0.598 | 1.0 (0.9–1.2) | 0.707 |
| rs7188750 | G/A | Intron 5 | Both | 18.5 | 0.226 | 1.0 (0.9–1.2) | 0.573 | 0.9 (0.8–1.1) | 0.354 | 1.2 (1.0–1.4) | 0.052 |
| rs8059139 | A/G | Intron 6 | Both | 11.1 | 0.326 | 1.0 (0.9–1.2) | 0.758 | 1.0 (0.8–1.2) | 0.627 | 1.1 (0.9–1.4) | 0.339 |
| rs3785076 | A/G | Intron 10 | Targeted | 11.7 | 0.327 | 1.0 (0.8–1.2) | 0.895 | 1.1 (0.8–1.3) | 0.684 | 0.9 (0.7–1.3) | 0.636 |
| rs4783689 | C/T | Intron 11 | Both | 32.9 | 0.250 | 1.0 (1.0–1.1) | 0.372 | 1.1 (1.0–1.2) | 0.134 | 1.0 (0.8–1.1) | 0.668 |
| rs16958383 | G/A | Intron 12 | Targeted | 20.7 | 0.542 | 1.0 (0.8–1.1) | 0.514 | 0.8 (0.7–1.0) | 0.020 | 1.3 (1.0–1.7) | 0.062 |
| rs10500545 | A/T | Intron 13 | Targeted | 12.6 | 0.989 | 1.0 (0.8–1.2) | 0.692 | 0.9 (0.7–1.1) | 0.194 | 1.1 (0.8–1.5) | 0.415 |
| rs9935563 | C/T | Intron 13 | Targeted | 35.7 | 0.620 | 1.0 (0.9–1.1) | 0.690 | 1.1 (0.9–1.3) | 0.213 | 0.8 (0.6–1.0) | 0.041 |
| rs9925080 | G/A | Intron 13 | Affy 6.0 | 9.3 | 0.182 | 1.0 (0.9–1.1) | 0.724 | 0.9 (0.7–1.1) | 0.281 | 1.1 (0.9–1.4) | 0.519 |
| rs9925161 | G/A | Intron 13 | Both | 9.3 | 0.111 | 1.0 (0.9–1.1) | 0.879 | 0.9 (0.7–1.1) | 0.304 | 1.1 (0.9–1.4) | 0.377 |
| rs8061932 | T/C | Intron 14 | Both | 22.8 | 0.855 | 1.0 (0.9–1.1) | 0.623 | 0.9 (0.8–1.0) | 0.073 | 1.1 (1.0–1.3) | 0.176 |
| rs3785078 | A/C | Intron 14 | Targeted | 13.7 | 0.585 | 0.9 (0.8–1.1) | 0.465 | 0.9 (0.7–1.1) | 0.223 | 1.1 (0.8–1.5) | 0.634 |
| rs9927789 | A/C | Intron 14 | Targeted | 19.0 | 0.214 | 1.0 (0.9–1.2) | 0.888 | 0.9 (0.8–1.1) | 0.565 | 1.1 (0.9–1.5) | 0.396 |
| rs7203904 | G/C | Intron 14 | Targeted | 32.5 | 0.706 | 1.0 (0.9–1.1) | 0.896 | 0.9 (0.8–1.0) | 0.128 | 1.2 (1.0–1.5) | 0.098 |
| rs2276329 | T/C | Intron 14 | Both | 9.7 | 0.257 | 1.0 (0.9–1.2) | 0.952 | 0.9 (0.8–1.1) | 0.582 | 1.1 (0.9–1.3) | 0.511 |
| rs13689 | T/C | 3′ UTR | Affy 6.0 | 19.0 | 0.397 | 1.0 (0.9–1.1) | 0.760 | 0.9 (0.8–1.1) | 0.220 | 1.2 (1.0–1.4) | 0.062 |
| rs17690554 | C/G | 3′ FRh | Affy 6.0 | 19.1 | 0.399 | 1.0 (0.9–1.2) | 0.834 | 0.9 (0.8–1.1) | 0.214 | 1.2 (1.0–1.4) | 0.082 |
| rs12447341 | C/T | 3′ FRh | Targeted | 29.0 | 0.698 | 1.0 (0.9–1.1) | 0.507 | 1.1 (0.9–1.3) | 0.474 | 0.8 (0.6–1.0) | 0.057 |
Major/minor alleles as determined by allele frequency among genotyped controls
Genotyping and study phase: Affymetrix targeted genotyping among 1,062 cases and 1,069 controls from phase 1 (Targeted), or Affymetrix 6.0 genotyping among 1,104 cases and 1,109 controls from Phase 1 and 969 cases and 975 controls from phase 2 (Affy 6.0), or genotyped by both (Both)
Minor allele freqency among genotyped controls
Hardy–Weinberg equilibrium test among controls
Risk of breast cancer per minor allele, adjusted for age, education, and study phase (when appropriate); A major allele, B minor allele; P value for trend
Premenopausal Women: 717 cases and 692 controls from phase 1 (Targeted), or 745 caes and 758 controls from phase 1 and 528 cases and 516 controls from phase 2 (Affy 6.0)
Postmenopausal Women: 345 cases and 377 controls from phase 1 (Targeted), or 369 caes and 419 controls from phase 1 and 441 cases and 459 controls from phase 2 (Affy 6.0)
3′ FR: 3′ flanking region, downstream of the CDH1 gene
Bold values considered to be significant P ≤ 0.05
As the etiology of breast cancer may differ by menopausal status, stratified analysis was conducted. Several CDH1 SNPs appeared to be associated with breast cancer risk among either pre- or postmenopausal women. Again, additive, dominant, and recessive models were considered, while for simplification, allelic associations are presented in Table 2. Polymorphisms of interest were then selected for further analysis to address whether associations with breast cancer risk were consistent when stratified by study phase (Table 3); models best suited to each SNP are presented. Among premenopausal women, three SNPs (rs2059254, rs9925923, and rs12919719) were consistently associated with increased risk in dominant models, whereas one SNP (rs7188750) was consistently associated with decreased risk in a recessive manner. Among postmenopausal women, four SNPs (rs9989407, rs7196495, rs7196661, and rs13689) were consistently associated with increased risk in recessive models, and five SNPs (rs2059254, rs9925923, rs12919719, rs12599393, and rs1862748) were consistently associated with decreased risk in a dominant manner. However, no associations retained statistical significance after adjusting for multiple comparisons. In order to further evaluate the hypothesis that the association between E-cadherin SNPs and breast cancer may differ in estrogen-related conditions, stratification by dichotomized BMI was also conducted; no associations were seen (data not shown).
Table 3.
Selected E-cadherin SNPs and breast cancer risk, by menopausal status and study phase, the Shanghai Breast Cancer Study
| SNP | Allelesa | Region | Modelb | Premenopausal womenc | |||||
|---|---|---|---|---|---|---|---|---|---|
| Phase 1 |
Phase 2 |
Combined |
|||||||
| OR (95% CI) | P Value | OR (95% CI) | P Value | OR (95% CI) | P Value | ||||
| rs9989407 | T/C | Promoter | R | 0.8 (0.5–1.2) | 0.286 | 1.1 (0.7–1.9) | 0.715 | 0.9 (0.7–1.3) | 0.576 |
| rs7196495 | T/C | Intron 2 | R | 0.8 (0.5–1.2) | 0.285 | 1.1 (0.7–1.9) | 0.630 | 0.9 (0.7–1.3) | 0.625 |
| rs7196661 | T/C | Intron 2 | R | 0.8 (0.6–1.2) | 0.345 | 1.1 (0.7–1.8) | 0.731 | 0.9 (0.7–1.3) | 0.636 |
| rs9941051 | T/C | Intron 2 | R | 0.8 (0.5–1.2) | 0.280 | 1.1 (0.7–1.9) | 0.630 | 0.9 (0.7–1.3) | 0.629 |
| rs2059254 | C/T | Intron 2 | D | 1.2 (1.0–1.6) | 0.073 | 1.3 (1.0–1.7) | 0.055 | 1.3 (1.1–1.5) | 0.009 |
| rs9925923 | C/T | Intron 2 | D | 1.3 (l.0–1.6) | 0.044 | 1.3 (1.0–1.7) | 0.053 | 1.3 (1.1–1.5) | 0.006 |
| rs12919719 | C/G | Intron 2 | D | 1.2 (0.9–1.5) | 0.133 | 1.3 (1.0–1.6) | 0.091 | 1.2 (1.0–1.5) | 0.027 |
| rs12599393 | C/T | Intron 2 | D | 1.2 (0.9–1.5) | 0.179 | 1.2 (0.9–1.5) | 0.211 | 1.2 (1.0–1.4) | 0.073 |
| rs1862748 | C/T | Intron 2 | D | 1.2 (0.9–1.5) | 0.206 | 1.2 (0.9–1.5) | 0.215 | 1.2 (1.0–1.4) | 0.080 |
| rs7188750 | G/A | Intron 5 | R | 0.4 (0.2–0.9) | 0.014 | 0.8 (0.4–1.5) | 0.427 | 0.6 (0.4–0.9) | 0.019 |
| rs13689 | T/C | 3′ UTR | R | 0.7 (0.4–1.1) | 0.143 | 0.7 (0.4–1.4) | 0.362 | 0.7 (0.5–1.1) | 0.086 |
| rs17690554 | C/G | 3′ FRe | R | 0.7 (0.4–1.1) | 0.142 | 0.7 (0.4–1.4) | 0.362 | 0.7 (0.5–1.1) | 0.085 |
| SNP | Allelesa | Region | Modelb | Postmenopausal womend | |||||
|---|---|---|---|---|---|---|---|---|---|
| Phase 1 |
Phase 2 |
Combined |
|||||||
| OR (95% CI) | P value | OR (95% CI) | P value | OR (95% CI) | P value | ||||
| rs9989407 | T/C | Promoter | R | 1.8 (1.0–3.2) | 0.041 | 1.3 (0.8–2.2) | 0.272 | 1.5 (1.0–2.2) | 0.031 |
| rs7196495 | T/C | Intron 2 | R | 1.8 (1.0–3.2) | 0.038 | 1.3 (0.8–2.2) | 0.305 | 1.5 (1.0–2.2) | 0.034 |
| rs7196661 | T/C | Intron 2 | R | 1.8 (1.0–3.2) | 0.037 | 1.3 (0.8–2.2) | 0.313 | 1.5 (1.0–2.2) | 0.035 |
| rs9941051 | T/C | Intron 2 | R | 1.8 (1.0–3.2) | 0.045 | 1.3 (0.8–2.1) | 0.362 | 1.5 (1.0–2.1) | 0.052 |
| rs2059254 | C/T | Intron 2 | D | 0.9 (0.6–1.3) | 0.536 | 0.7 (0.5–1.0) | 0.026 | 0.8 (0.6–1.0) | 0.032 |
| rs9925923 | C/T | Intron 2 | D | 0.9 (0.6–1.2) | 0.393 | 0.7 (0.6–1.0) | 0.049 | 0.8 (0.6–1.0) | 0.034 |
| rs12919719 | C/G | Intron 2 | D | 0.9 (0.6–1.2) | 0.439 | 0.7 (0.5–1.0) | 0.029 | 0.8 (0.6–1.0) | 0.025 |
| rs12599393 | C/T | Intron 2 | D | 0.9 (0.6–1.2) | 0.340 | 0.7 (0.5–1.0) | 0.031 | 0.8 (0.6–1.0) | 0.016 |
| rs1862748 | C/T | Intron 2 | D | 0.9 (0.6–1.2) | 0.376 | 0.7 (0.5–0.9) | 0.019 | 0.8 (0.6–0.9) | 0.014 |
| rs7188750 | G/A | Intron 5 | R | 1.6 (0.7–3.7) | 0.256 | 1.6(0.8–3.0) | 0.195 | 1.6 (1.0–2.7) | 0.070 |
| rs13689 | T/C | 3′ UTR | R | 1.6 (0.8–3.2) | 0.183 | 1.7 (0.8–3.4) | 0.158 | 1.7 (1.0–2.7) | 0.043 |
| rs17690554 | C/G | 3′ FRe | R | 1.5 (0.8–3.0) | 0.234 | 1.7 (0.8–3.4) | 0.158 | 1.6 (1.0–2.6) | 0.059 |
Major/minor alleles as determined by allele frequency among genotyped controls
Model of effect: dominant (D): AB and BB versus AA, or recessive (R): BB versus AA and AB; AA major allele homozygotes, AB heterozygotes, BB minor allele homozygotes
Risk of breast cancer, adjusted for age and education for premenopausal women; Phase 1 includes 745 cases and 758 controls, Phase 2 includes 528 cases and 516 controls
Risk of breast cancer, adjusted for age and education for postmenopausal women; Phase 1 includes 369 cases and 419 controls, Phase 2 includes 441 cases and 459 controls
3′ FR: 3′ flanking region, downstream of the CDH1 gene
Bold values considered to be significant P ≤ 0.05
The LD structure of the 40 polymorphic E-cadherin SNPs evaluated in the current study is shown in Fig. 1. This LD structure included 2,152 genotyped controls and contained seven haplotype blocks. No associations with breast cancer risk were found in analyses among all women. However, E-cadherin haplotypes and breast cancer risk seemed to be associated in analysis that included only either pre- or postmenopausal women (data not shown). In general, haplotype analysis was consistent with the results from single SNP analysis.
Fig. 1.
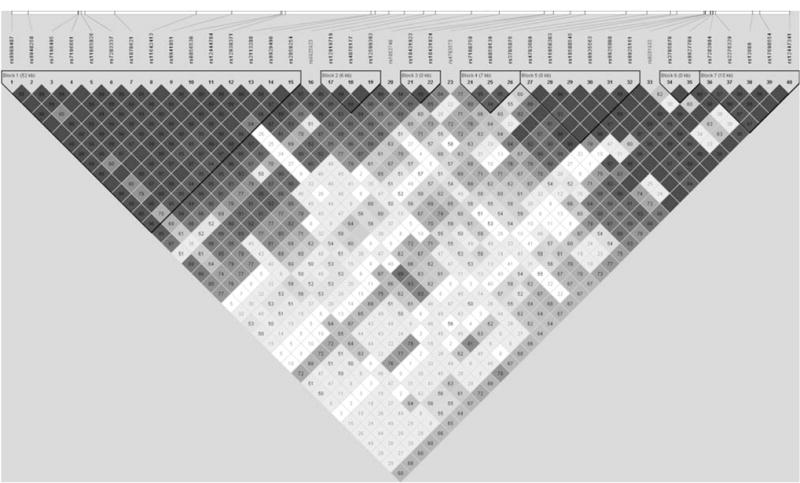
Linkage disequilibrium (LD) structure of 40 E-cadherin polymorphisms among 2,151 controls from the Shanghai Breast Cancer Study, value shown is D′
Discussion
Common genetic variation across the E-cadherin gene was systematically evaluated in a large, population-based study of Chinese women. A total of 46 SNPs were genotyped; no overall associations with breast cancer risk were observed among 2,083 cases and 2,152 controls. In addition to polymorphisms, many other common CDH1 alterations have been reported, including mutations, loss of heterozygosity, transcriptional repression, and epigenetic silencing [3]. If present, these mechanisms of E-cadherin loss could dilute any effects due to SNPs on breast cancer risk, possibly explaining our results. However, several associations with breast cancer risk were observed when the study population was stratified by menopausal status; many were consistent between study phases. Differences in the relationship between CDH1 polymorphisms and breast cancer risk by menopausal status could result from the complex interaction between E-cadherin and the estrogen pathway [21].
A classical tumor suppressor, E-cadherin expression has been shown to be frequently reduced or lost among epithelial tumors [2, 3]. This results in the suboptimal regulation of cell–cell adhesion, loss of cellular polarity, tissue disorganization, tumor progression, and metastasis [1, 3]. Roles in tumor initiation have also been suggested, as the loss of E-cadherin may promote tumorigenesis by releasing membrane-bound β-catenin, thereby, potentiating the canonical Wnt signaling pathway, or by modulating mitogenic signaling, such as EGF-induced cellular proliferation [1]. Several functional polymorphisms that diminish E-cadherin expression have been reported [4, 6, 10, 11]; however, studies on breast cancer risk have been sparse. Yu et al. genotyped the functional promoter polymorphisms −160 C/A (rs16260) and −347 G/GA among 468 cases and 470 controls and found that the two SNPs were in high LD [9]. They found a significant dominant effect, such that minor allele carriers (rs16260 A) were 30% more likely to be breast cancer cases than women with only the major allele (C) [9]. On the contrary, Lei et al. genotyped the −160 C/A (rs16260) SNP among 576 cases and 348 controls, and found no association with breast cancer risk [5]. While not directly genotyped in this study, the genetic variation of this polymorphism was captured; three genotyped SNPs (rs11865026, rs8056538, and rs12930371) are reported to be in perfect LD (D′ = 1.0, r2 = 1.0) with rs16260 [17]. None of these three SNPs were associated with breast cancer risk, either among all women, or when stratified by menopausal status, in this study.
Several E-cadherin polymorphisms were found to be associated with breast cancer risk in analyses stratified by menopausal status, and significant when data from both study phases were combined. Among premenopausal women, a modest increase in risk was associated with three SNPs in intron 2, while a larger protective effect was observed for an SNP in intron 5. Among postmenopausal women, a large increase in risk was seen for SNPs in the promoter, intron 2, and 3′ UTR, while a modest decrease in risk was associated with five SNPs in intron 2. However, several considerations must be made in interpreting these results. First, it must be noted that the sample size of this stratified analysis was reduced, and the smallest number of participants that was included was for those SNPs assayed by Affymetrix Targeted Genotyping among postmenopausal women (345 cases and 377 controls). In addition, when considering the number of associations evaluated, a Bonferroni corrected P value of 0.00125 would replace the 0.05 threshold for statistical significance; none of our estimates met this level of significance.
Strengths of this study include a large, two-phase, population-based study. We also had excellent coverage of the genetic variation across E-cadherin, as the polymorphisms that we genotyped are estimated to cover 100% of the SNPs with MAFs of 5% or greater with an r2 of 0.8. Further, we had greater than 89% power to detect the association of 1.3 or greater for an SNP with an MAF of 15%, should such an association exist. In summary, several CDH1 SNPs were found to be differentially associated with pre- and postmenopausal women, with consistent results between our two study phases. Additional studies are warranted to evaluate the association between E-cadherin polymorphisms and breast cancer susceptibility.
Acknowledgments
This research was supported by USPHS grants R01CA64277, R01CA90899, and R01CA124558. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health. The authors wish to thank the participants and research staff of the Shanghai Breast Cancer Study for their contributions and commitment to this project, and Brandy Venuti for assistance with the preparation of this manuscript. Sample preparation and genotyping assays, using Affymetrix arrays, were conducted at the Survey and Biospecimen Shared Resource and the Vanderbilt Microarray Shared Resource, respectively, which are supported in part by the Vanderbilt-Ingram Cancer Center (P30CA68485).
References
- 1.Jeanes A, Gottardi CJ, Yap AS. Cadherins and cancer: how does cadherin dysfunction promote tumor progression? Oncogene. 2008;27:6920–6929. doi: 10.1038/onc.2008.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van RF, Berx G. The cell–cell adhesion molecule E-cadherin. Cell Mol Life Sci. 2008;65:3756–3788. doi: 10.1007/s00018-008-8281-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baranwal S, Alahari SK. Molecular mechanisms controlling E-cadherin expression in breast cancer. Biochem Biophys Res Commun. 2009;384:6–11. doi: 10.1016/j.bbrc.2009.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li LC, Chui RM, Sasaki M, Nakajima K, Perinchery G, Au HC, et al. A single nucleotide polymorphism in the E-cadherin gene promoter alters transcriptional activities. Cancer Res. 2000;60:873–876. [PubMed] [Google Scholar]
- 5.Lei H, Sjoberg-Margolin S, Salahshor S, Werelius B, Jandakova E, Hemminki K, et al. CDH1 mutations are present in both ductal and lobular breast cancer, but promoter allelic variants show no detectable breast cancer risk. Int J Cancer. 2002;98:199–204. doi: 10.1002/ijc.10176. [DOI] [PubMed] [Google Scholar]
- 6.Cattaneo F, Venesio T, Molatore S, Russo A, Fiocca R, Frattini M, et al. Functional analysis and case–control study of −160C/A polymorphism in the E-cadherin gene promoter: association with cancer risk. Anticancer Res. 2006;26:4627–4632. [PubMed] [Google Scholar]
- 7.Wang GY, Lu CQ, Zhang RM, Hu XH, Luo ZW. The E-cadherin gene polymorphism 160C→A and cancer risk: A HuGE review and meta-analysis of 26 case–control studies. Am J Epidemiol. 2008;167:7–14. doi: 10.1093/aje/kwm264. [DOI] [PubMed] [Google Scholar]
- 8.Qiu LX, Li RT, Zhang JB, Zhong WZ, Bai JL, Liu BR, et al. The E-cadherin (CDH1)−160 C/A polymorphism and prostate cancer risk: a meta-analysis. Eur J Hum Genet. 2009;17:244–249. doi: 10.1038/ejhg.2008.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu JC, Hsu HM, Chen ST, Hsu GC, Huang CS, Hou MF, et al. Breast cancer risk associated with genotypic polymorphism of the genes involved in the estrogen-receptor-signaling pathway: a multigenic study on cancer susceptibility. J Biomed Sci. 2006;13:419–432. doi: 10.1007/s11373-006-9069-7. [DOI] [PubMed] [Google Scholar]
- 10.Nakamura A, Shimazaki T, Kaneko K, Shibata M, Matsumura T, Nagai M, et al. Characterization of DNA polymorphisms in the E-cadherin gene (CDH1) promoter region. Mutat Res. 2002;502:19–24. doi: 10.1016/s0027-5107(02)00024-6. [DOI] [PubMed] [Google Scholar]
- 11.Shin Y, Kim IJ, Kang HC, Park JH, Park HR, Park HW, et al. The E-cadherin −347G→GA promoter polymorphism and its effect on transcriptional regulation. Carcinogenesis. 2004;25:895–899. doi: 10.1093/carcin/bgh073. [DOI] [PubMed] [Google Scholar]
- 12.Tsai FJ, Wu HC, Chen HY, Lu HF, Hsu CD, Chen WC. Association of E-cadherin gene 3′-UTR C/T polymorphism with calcium oxalate stone disease. Urol Int. 2003;70:278–281. doi: 10.1159/000070135. [DOI] [PubMed] [Google Scholar]
- 13.Nasri S, More H, Graziano F, Ruzzo A, Wilson E, Dunbier A, et al. A novel diffuse gastric cancer susceptibility variant in E-cadherin (CDH1) intron 2: a case control study in an Italian population. BMC Cancer. 2008;8:138. doi: 10.1186/1471-2407-8-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao YT, Shu XO, Dai Q, Potter JD, Brinton LA, Wen W, et al. Association of menstrual and reproductive factors with breast cancer risk: results from the Shanghai Breast Cancer Study. Int J Cancer. 2000;87:295–300. doi: 10.1002/1097-0215(20000715)87:2<295::aid-ijc23>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 15.Beeghly-Fadiel A, Long JR, Gao YT, Li C, Qu S, Cai Q, et al. Common MMP-7 Polymorphisms and Breast Cancer Susceptibility: A Multistage Study of Association and Functionality. Cancer Res. 2008;68:6453–6459. doi: 10.1158/0008-5472.CAN-08-0636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng W, Long J, Gao YT, Li C, Zheng Y, Xiang YB, et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat Genet. 2009;41:324–328. doi: 10.1038/ng.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 18.de Bakker PIW, McVean G, Sabeti PC, Miretti MM, Green T, Marchini J, et al. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat Genet. 2006;38:1166–1172. doi: 10.1038/ng1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 20.Lin DY, Zeng D, Millikan R. Maximum likelihood estimation of haplotype effects and haplotype-environment interactions in association studies. Genet Epidemiol. 2005;29:299–312. doi: 10.1002/gepi.20098. [DOI] [PubMed] [Google Scholar]
- 21.Oesterreich S, Deng W, Jiang S, Cui X, Ivanova M, Schiff R, et al. Estrogen-mediated down-regulation of E-cadherin in breast cancer cells. Cancer Res. 2003;63:5203–5208. [PubMed] [Google Scholar]


