FIG 5.
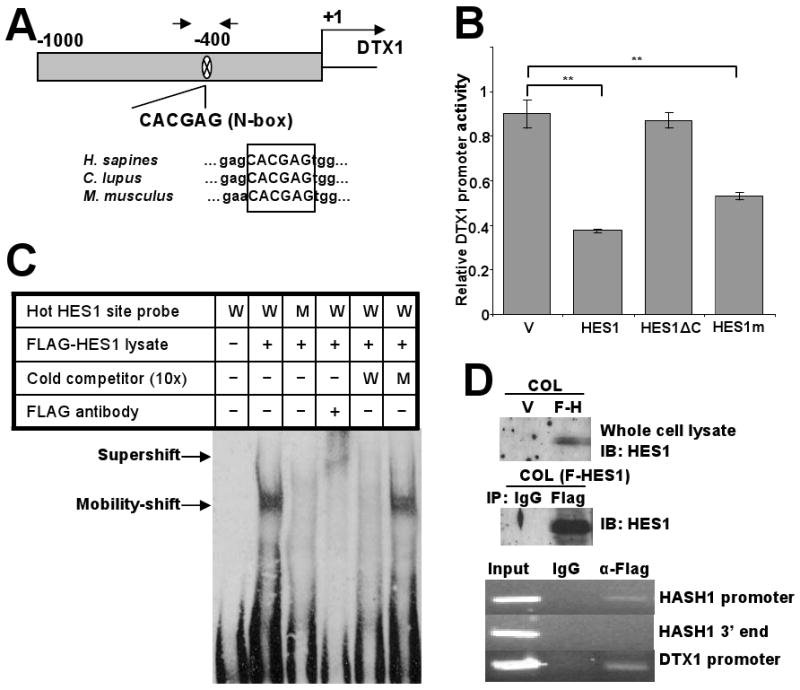
HES1 actively represses the DTX1 through binding to an N-box motif in the DTX1 promoter. (A) Schematic depiction of the 1kb DTX1 proximal promoter in the 5′ untranslated region. One potential HES1 binding site (N-box) was located at -400bp. Arrows represent the position of the PCR primers utilized for the ChIP analysis of the following panels (upper part). Sequence alignment of the DTX1 genes of human, dog and mouse showed the identical N-box in the similar position (lower part). (B) DTX1 promoter reporter (Renilla luciferase) was cotransfected with a firefly luciferase control into 293T cells transiently expressing vector control, full length HES1, a mutant HES1 with the C-terminus deleted (HES1ΔC) or HES1-WRPW mutant (HES1m), which is predicted to have less Groucho/TLE binding. Relative DTX1 promoter activity was determined after 48 hours. (**, p<0.005). (C) Gel mobility-shift analysis showing HES1 binding to the N-box motif in the DTX1 promoter. Nuclear protein extract (3μg) from 293T cells expressing FLAG-HES1 were incubated with biotin labeled probes containing WT N-box (W), mutant N-box (M), and anti-FLAG antibody, either alone or in combination, at room temperature for 15 min. As cold competitors, excess doses (10-fold) of non-biotinylated WT or mutant oligomers were included. Arrows indicate the specific N-box DNA-HES1 complex and the HES1 supershifted complex. (D) Chromatin immunoprecipitation (ChIP) assay: COL cells were transduced with FLAG-tagged HES1. Immunoblot (IB) confirmed expression of HES1 and effective immunoprecipitation (IP) of HES1 with anti-FLAG antibody (upper panels). Sheared chromatin from transfected cells was immunoprecipitated with anti-FLAG antibody or nonspecific IgG as control. Immunoprecipitated DNA was used as template in PCR for HASH1 promoter region (positive control), HASH1 3′ end region (negative control) or DTX1 promoter region (lower panels).
