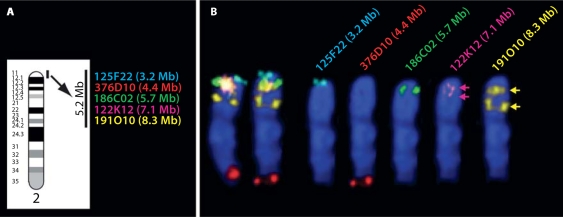
Fig. 3.
FISH analysis of five BAC clones from the CFA2qcen region of the genome assembly. The five most proximal clones selected for analysis in phase I are shown in A against the ideogram of CFA2, spanning a 5.2 Mb region of the genome assembly. (B) Left, the composite results of FISH analysis of these clones on both CFA2 homologues from one cell and right, the five separate planes of fluorescent signal corresponding to each differentially labeled clone are shown individually for one of these homologues. Two clones (125F22 and 186C02) showed unique signal in the expected chromosome location. Clones 122K12 and 191O10 also mapped where expected, but showed additional hybridization sites further down the chromosome, which were of equal strength to the primary signal. These pairs of probe signals (arrowed) suggested the presence of tandem repeat sequences in this region of CFA2, which was supported by the identification of a similar pattern of results for the next distal marker from this region (199D17, CFA2; 8.9 Mb, data not shown). Clones distal to this marker mapped to the expected unique location, thus the region of repetitive sequence appears to extend to approximately 9 Mb from the start of the CFA2 assembly. Note that clone 376D10 (CFA2; 4.3 Mb according to the assembly) mapped significantly more distally than expected, close to the CFA2 telomere. Through co-hybridization of 376D10 with clones from this region (following the strategy outlined in Fig. 2), the cytogenetic location of this BAC was resolved to approximately CFA2; 87 Mb. This finding was subsequently attributed to misplacement of one end of the BAC clone sequence at CFA2; 4.3 Mb, most likely due to the presence of repetitive sequence, whilst the other end was correctly positioned at CFA2; 86.7 Mb.