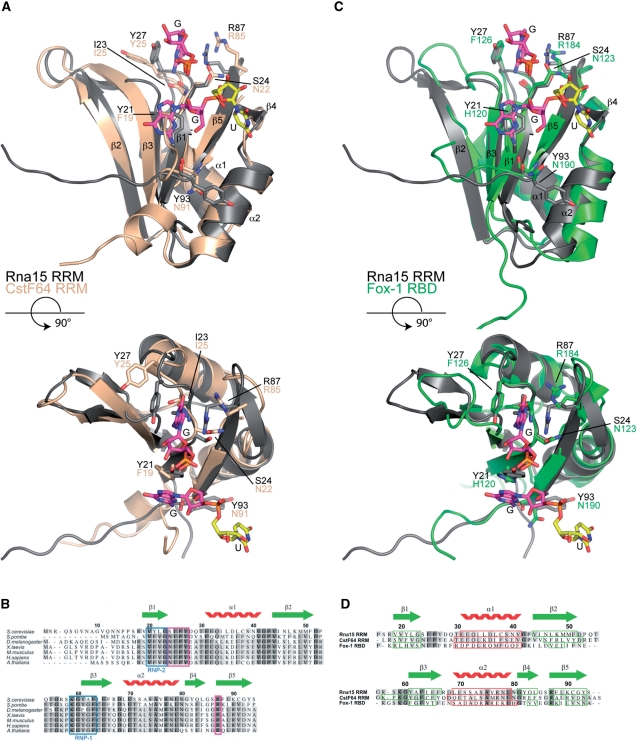
Figure 7.
Sequence and structure based alignments. (A) Structural superposition of the Rna15(16-111)-GUUGU complex onto the RRM of CstF64 (pdb code:1P1T), (r.m.s.d. Rna15 residues 16-94 is 1.25Å over 77 Cα positions, 44% sequence identity). Rna15(16-111) is shown in grey, CstF64 is coloured wheat. A GU dinucleotide has been modeled at Site-II by superposition of the Rna15(16-111)-GUUGU and Rna15(16-111)ht-GUUGU structures. Bound ribonucleotides and residues that mediate protein–RNA interactions are shown as sticks, secondary structure elements and termini are labelled. (B) Multiple primary sequence alignment of Rna15 RRM, species range from S. cerevisiae to A. thaliana. The RNP-1 and RNP-2 motifs are highlighted in blue, residues that form the Site-I base binding pocket are boxed in magenta and the Rna15 RRM secondary structure elements are shown above. (C) Structural superposition of the Rna15(16-111)-GUUGU complex onto the RRM of Fox-1 (pdb code:2ERR), (r.m.s.d. Rna15 residues 16-94 is 1.55Å over 73 Cα positions, 19.2% sequence identity) Rna15(16-111) and the Fox-1 RRM are shown in grey and green respectively. Bound nucleotides and residues involved in protein–RNA interactions are highlighted as in (A). (D) Structure based sequence alignment of the RRMs from Rna15, CstF64 and Fox-1. Rna15 sequence numbering and secondary structure elements are shown above the aligned sequences. Darker to lighter grey depicts greater to lower sequence conservation, secondary structure elements are boxed in all sequences, β-strands in green and α-helices in red.