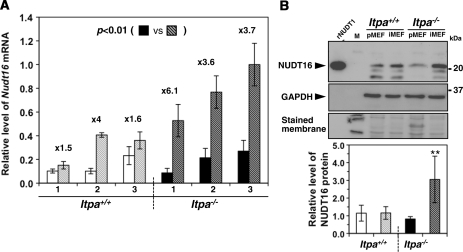
Figure 6.
NUDT16 with strong dIDP/IDP-hydrolyzing activity is the back-up enzyme responsible for the cancellation of ITPA-deficient phenotypes during immortalization. (A) Expression of Nudt16 mRNA. Quantitative real-time RT–PCR was performed to compare Nudt16 mRNA levels between primary (passage 3) and immortalized MEFs. Levels of Nudt16 mRNA were normalized to those of Gapdh mRNA. Values relative to the highest level of Nudt16 mRNA in immortalized Itpa−/– MEFs (isolate No. 3) are shown. Unpaired Student’s t-test (two-tailed) showed P < 0.01 between primary and immortalized Itpa−/– MEFs. Data are shown in a bar graph (mean ± SD), with fold changes between primary and immortalized MEFs (n = 3 independent isolates). Open and black bars show primary MEFs; shaded bars indicate immortalized MEFs. (B) Expression of NUDT16 protein. Western blotting was performed for crude cell extracts (40 µg) prepared from primary (Passage 2) and immortalized MEFs, using anti-NUDT16 antibody (top). GAPDH was detected as an internal control (middle). Membranes were stained with Gel Code Blue in order to confirm the loading and transfer (bottom). Intensities of NUDT16 bands were measured and relative levels normalized to GAPDH are shown in a bar graph. Open and black bars show primary MEFs; shaded bars indicate immortalized MEFs. Result of non-repeated measures ANOVA (two-tailed), P < 0.019. Bonferroni post hoc test, **P < 0.01 (versus others). Error bars represent the SD (n = 3 independent isolates).