Abstract
In this study we examined whether established signal transduction cascades, p44/42 mitogen activated protein kinase (ERK1/2) and Jun N-terminal kinases (JNK) pathways, are altered in N2a neural cells in response to proteasome inhibition. Additionally, we sought to elucidate the relative contribution of these signal transduction pathways to the multiple downstream effects of proteasome inhibition. Our data indicate that ERK1/2 and JNK are activated in response to proteasome inhibition. Washout of proteasome inhibitor (MG132) results in an enhancement of ERK1/2 activation and amelioration of JNK activation. Treatment with an established MAPK inhibitor resulted in an increase in proteasome inhibitor toxicity, while incubation with JNK inhibitor was observed to significantly attenuate proteasome inhibitor toxicity. Subsequent studies demonstrate that inhibition of ERK1/2 and JNK activity does not alter the gross increase in ubiquitinated protein following proteasome inhibitor administration. Similarly, ERK1/2 and JNK activity do not appear to play a role in the disruption of polysomes following proteasome inhibitor administration in neural cells. Together, these data indicate that ERK1/2 and JNK activation may play differential roles in modulating neurochemical disturbances and neurotoxicity induced by proteasome inhibition.
Keywords: Aging, neuron, neurotoxicity, protein degradation, protein synthesis, signal transduction
INTRODUCTION
The proteasome is a large protein complex which is responsible for a significant amount of overall intracellular proteolysis, including the degradation of the majority of short lived proteins (Shringarpure et al., 2002; Goldberg, 2003). Proteasome inhibition occurs during aging and in a variety of age-related neurodegenerative conditions (Chondrogianni et al., 2005; Keller et al., 2002), and is believed to contribute to multiple aspects of neuropathology and neurotoxicity. The majority of studies to date have focused on the role of proteasome inhibition as a mediator of increased ubiquitin-protein conjugates and protein aggregation within a variety of cell types and tissues (Chondrogianni et al., 2003; Sullivan et al., 2004; Rideout et al., 2001, 2003; Hyun et al., 2003; Li et al., 2008). More recent studies have demonstrated a role for proteasome inhibition as a mediator of decreased protein synthesis (Ding et al., 2006), and a mediator of ribosome dysfunction (Ding et al., 2006; Kim et al., 2005; Jiang and Wek, 2005; Othumpangat et al., 2005), with such disruptions potentially contributing to the toxicity of proteasome inhibition. Such studies not only identify interplay between protein synthesis and protein degradation, but also open the possibility of proteasome inhibition contributing to cytotoxicity through modulation of protein synthesis (Ding et al., 2007).
A number of signal transduction cascades have been demonstrated to be modulated in response to proteasome inhibition, including the p44/42 mitogen activated protein kinase (ERK1/2) and Jun N-terminal kinases (JNK) pathways (Shi et al., 2006; Yamamoto et al., 2008; Li et al., 2008; Fineschi et al., 2008; Liu et al., 2008; Lam and Cadenas, 2008). The regulation of these signal transduction cascades by stressors such as proteasome inhibition appears to be extremely cell type specific, with the corresponding effects of these signal transduction pathways on cellular homeostasis also being extremely cell type specific. For example, studies have demonstrated both pro-apoptotic and anti-apoptotic roles for ERK1/2 and JNK activation (Junttila et al., 2008; Borsello and Forloni, 2007; Raman et al., 2007). Interestingly, proteasome inhibition itself has been demonstrated to be both pro- and anti-apoptotic in a cell type specific manner (Meiners et al., 2008; Montagut et al., 2006; Vu et al., 2008; Sun et al., 2008; Harris et al., 2008). Exploring the potential role of ERK1/2 and JNK activation in modulating the toxicity of proteasome inhibition is therefore an extremely important and relevant topic to aging and age-related diseases of the brain, where proteasome inhibition is known to occur.
Recent studies have demonstrated that the toxicity of proteasome inhibition in rat primary neurons is reversible (Ding et al., 2006), where washout of the proteasome inhibitor during the first 12 hours of treatment results in a significant attenuation in neural death (Ding et al., 2006). In the present study we sought to utilize this model to elucidate the relationship between the reversible effects of proteasome inhibition as related to alterations in signal transduction, ubiquitinated protein levels, ribosome alterations, and neural viability. Together, these data indicate potentially different roles for ERK1/2 and JNK in regulating the toxicity of proteasome inhibition in neural cells, and indicate that the relationship between ERK1/2 and JNK with proteasome inhibitor toxicity is possibly independent of effects on ubiquitinated protein levels or effects on ribosome homeostasis.
MATERIALS AND METHODS
Materials
All cell culture supplies were obtained from GIBCO Life Sciences (Gaithersburg, MD, USA). Proteasome inhibitor MG132, ERK1/2 inhibitor 3-(2-Aminoethyl)-5-((4-ethoxyphenyl)methylene)-2,4-thiazolidinedione·HCl (catalog # 328006), JNK inhibitor AEG3482 (catalog # 152228) were obtained from Calbiochem (San Diego, CA). Antibodies against p44/42 ERK1/2 (catalog # 4695), phosphorylated p44/42 ERK1/2 (catalog # 9101S), JNK (catalog # 9258), and phosphorylated JNK (catalog # 4668S) were purchased from Cell Signaling Technology (Danvers, MA); antibodie anti-β-actin (sc-47778) were purchased from Santa Cruz Biotech, Inc. (Santa Cruz, CA). The secondary antibodies peroxidase-conjugated goat anti-rabbit IgG (H+L) (catalog # 111-035-003) and peroxidase-conjugated goat anti-mouse IgG (H+L) (catalog # 115-035-003) were purchased from Jackson ImmunoResearch Lab, Inc. (West Grove, PA). Protease inhibitor was purchase from Roche Diagnostics (Indianapolis, IN); phosphotase inhibitor cocktail 1 (catalog # P2850) and phosphotase inhibitor cocktail 2 (catalog # P5726) were purchased from Sigma (St. Louis, MO). All Western-blot supplies, 7.5% precast gel (catalog # 161–1154) and 10–20% precast gel (catalog # 161–1160) were purchased from BIO-RAD (Hercules, CA). Polysome analyzing system was purchased from Brandel (Gaithersburg, MD). All other chemicals were purchased from Sigma (St. Louis, MO).
Cell Culture
N2a cells were cultured in 5% CO2 at 37°C in minimum essential medium (MEM) containing 5% fetal bovine serum, 1% of antibiotic antimycotic and 1 mM pyruvate. All cells were plated in fresh medium one day prior to experimentation and were used at ~70% confluency.
Western blot analysis
N2a cells were lysed in lysis buffer (50 mM Tris-HCl, pH 8.0, 20% glycerol, 140 mM NaCl, 0.5% Nonidet P-40, 5 mM MgCl2, 0.2 mM EDTA) with protease inhibitors and phsophotase inhibitors. Proteins were electrophoresed and blotted to nitrocellulose membrane. Blots were probed with antibodies and visualized with peroxidase-linked secondary antibodies by using Pierce ECL Western blotting substract (Pierce, Rockford, IL).
Analysis of cell survival
Cell survival was determined by quantification of apoptotic and necrotic nuclei using Hoechts 344 staining as described previously (Keller et al., 1998). Briefly, N2a cells were stained with the fluorescent DNA-binding dye Hoechts 344 (Kruman et al., 1997), and the percentage of viable cells determined by counting the number of dead cells (condensed and fragmented nuclei) using a fluorescence microscope equipped with a 32X objective. Additional confirmation of cell viability was determined using MTT reduction as a measure of cell viability as reported previously (Keller et al., 1998), data not shown.
Analysis of ribosomes and polysome levels
Ribosomes and polyribosome fractions were purified and quantified as described in previous studies from our laboratories and by others (Ding et al., 2006; Zhou et al., 2008). Briefly, N2a cells were cultured in Dulbecco’s modified Eagle’s medium, as highlighted above, in the presence of 10 μM MG132, or to no MG132, for 6 h. At that time 10 μg/ml cycloheximide was added to the medium prior to collection and analysis. Cells were washed in cold phosphate-buffered-saline solution supplemented with 10 μg/ml cycloheximide, and then lysed with ice-cold lysis buffer containing 20 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 100 mM NaCl, 0.4% Nonidet P-40, and 10 μg/ml cycloheximide. The extracts were passed through a 23-gauge needle for proper lysis of cells, incubated for 10 min on ice, and insoluble material was collected by microcentrifugation at 10,000 rpm for 10 min at 4 °C. The resulting supernatant was then applied onto a 15–50% sucrose gradient containing 20 mM Tris-HCl (pH 7.5), 5 mM MgCl2, 100 mM NaCl, 10 μg/ml cycloheximide, and centrifuged for 2 h at 40,000 rpm in a Beckman SW-41Ti rotor. Following centrifugation, the gradients were fractionated by Brandel fractionation system, and the absorbance of cytosolic RNA at 254 nm calculated using an in-line UV monitor.
RESULTS
Reversible toxicity of proteasome inhibitors in N2a cells
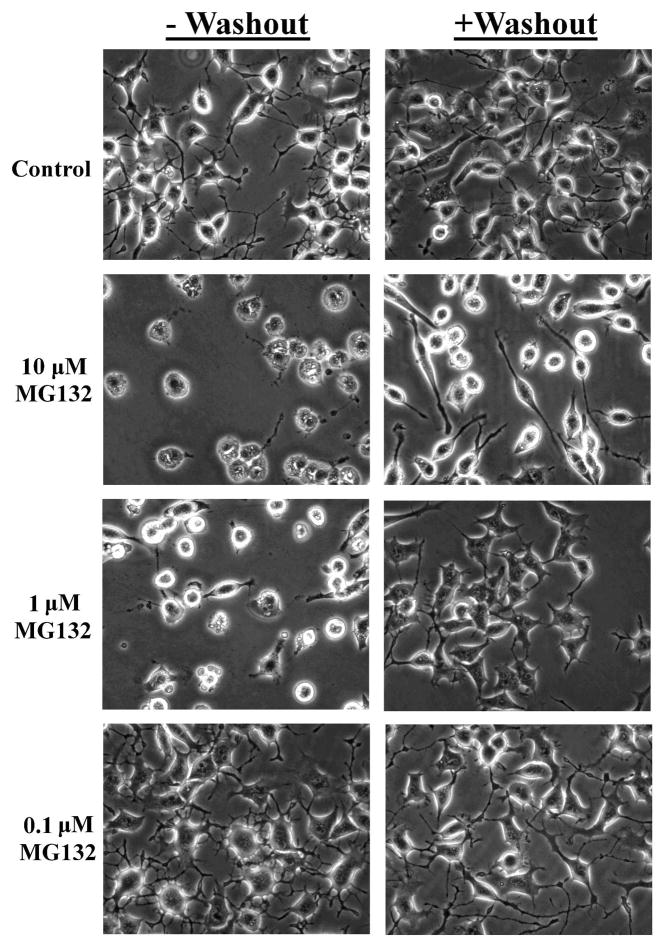
Previously we have demonstrated that the ability of proteasome inhibitors to promote reversible toxicity in primary neurons (Ding et al., 2006), whereby the washout of the proteasome inhibitor within 12 hours of administration ameliorates neurotoxicity. Such studies suggest that the event(s) which mediate neurotoxicity must be activated to a specific degree, or in concert with other neurochemical changes, in order to induce neuron death. In the present study we examined whether proteasome inhibitor toxicity is similarly reversible in N2a neural cells. In these studies N2a cells were treated with increasing concentrations of the reversible proteasome inhibitor MG132. Analysis of neural survival demonstrated that there was a dose-dependent increase in toxicity that was largely reversible by washout of the proteasome inhibitor (Fig. 1 & 2). These data indicate that N2a cells respond similarly to rat primary neurons, in regards to neurotoxicity following proteasome inhibitor treatment.
Figure 1. Analysis of MG132 toxicity in N2a neural cells.
Increasing concentrations of MG132 increase cytotoxicity as evidenced by a loss of cell integrity and blebbing, with washout of proteasome inhibitor largely preserving cell viability. Examples of cell morphology are presented following 24 hrs of treatment with increasing concentrations of the proteasome inhibitor MG132 (-washout). Additionally, cultures which received a 3 hr treatment with MG132, followed by washout of the inhibitor and quantification of viability 24 hrs following proteasome inhibitor administration are presented (+ washout).
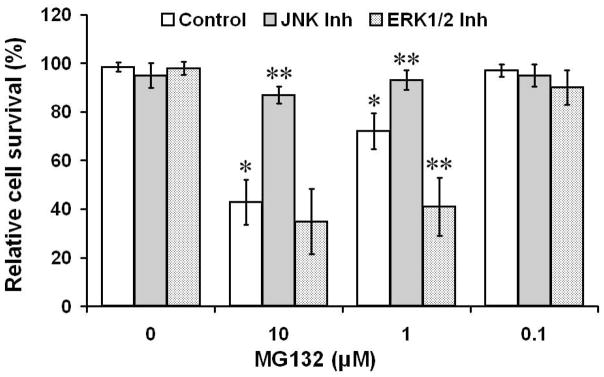
Figure 2. Analysis of MG132 toxicity in neural N2a cells.
Increasing concentrations of MG132 were administered to neural N2a cells with the cells analyzed for neural viability 24 hours following initial MG132 treatment. In each experimental group (0–10 μM MG132) neural cells were also treated with 5 μM ERK1/2 inhibitor or 5 μM JNK inhibitor (AEG1842). *p < 0.05 compared to cells not treated with MG132; **p < 0.05 compared to cells not treated with ERK1/2 or JNK inhibitors
Effect of proteasome inhibitors on ERK1/2 and JNK activation
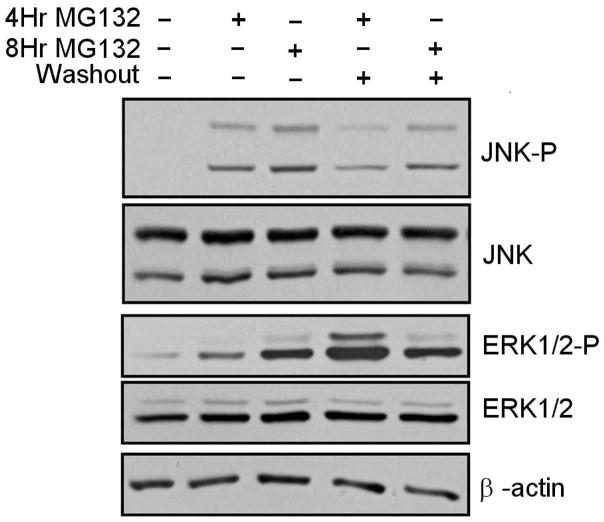
In order to determine whether proteasome inhibitors modulate the activation of the ERK1/2 and JNK pathways in N2a cells, we conducted studies in which cells were treated with MG132, or treated with MG132 followed by washout of the inhibitor. In these studies we observed that the presence of MG132 promoted a modest elevation in ERK1/2 activation, as indicated by the presence of increased detection of phosphorylated ERK1/2, which is known to be an activated form of ERK1/2 (Fig. 3). Interestingly, the greatest activation of ERK1/2 was observed to occur during the washout of MG132 (Fig. 3), which was also associated with neural survival (Fig. 1 & 2). We next examined the degree to which MG132 was able to modulate JNK activation, using phosphorylated JNK levels as an index of JNK activation. In these studies we observed that JNK appeared to be activated in response to MG132, with washout of MG132 ameliorating the levels of JNK activation (Fig. 3).
Figure 3. Analysis of ERK1/2 and JNK activation following treatment with proteasome inhibitor MG132.
Increased levels of ERK1/2 and JNK activation are observed following proteasome inhibitor treatment, with washout of proteasome inhibitor promoting further ERK1/2 activation and ameliorating the elevation in JNK activation. The levels of activated p44/42 MAP kinase (P-ERK1/2), non-phosphorylated ERK1/2 (ERK1/2), activated JUN kinase (P-JNK) and non-phosphorylated Jun Kinase (JNK) were analyzed following 4 or 8 hours of 10 μM MG132 treatment. Additionally, cultures were analyzed following washout of MG132 and incubation of cells in complete N2a media (lacking MG132) for 16 hours. Data representative of results from 3 separate experiments.
Effect of ERK1/2 and JNK inhibitors on proteasome inhibitor-induced toxicity
In order to begin to elucidate the downstream consequences of ERK1/2 and JNK activation to proteasome inhibitor-induced neurotoxicity, we treated N2a cells with ERK1/2 and JNK inhibitors during the different experimental paradigms involving MG132 treatment. In these studies we observed that addition of a ERK1/2 inhibitor did not reduce MG132 toxicity and actually promoted an exacerbation of MG132 toxicity (Fig 2). In contrast, treatment with JNK inhibitor resulted in an amelioration of MG132 toxicity (Fig 2). Together, these data indicate that MAP kinase and JNK pathways appear to play differential roles in modulating neural viability in response to proteasome inhibition.
Effects of ERK1/2 and JNK inhibition on MG132-induced increases in ubiquitinated protein
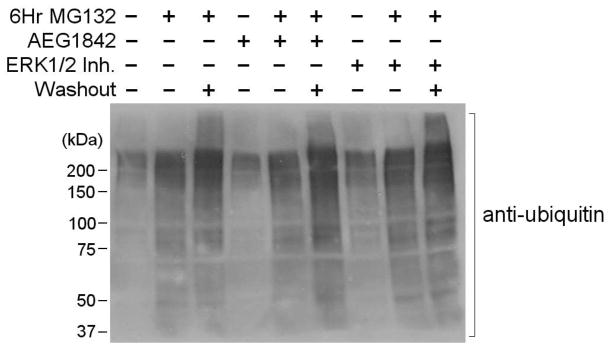
We next examined the potential roles of ERK1/2 and JNK activation to MG132-induced increases in ubiquitinated protein. In these studies we observed that there was a predicted increase in ubiquitinated protein levels upon treatment with MG132, which was sustained in response to washout of the proteasome inhibitor (Fig. 4). Administration of ERK1/2 and JNK inhibitors did not significantly alter the level of ubiquitinated protein during MG132 treatment or following washout of the proteasome inhibitor (Fig. 4). These studies suggest that ERK1/2 and JNK do not significantly modulate the gross levels of ubiquitinated protein following proteasome inhibition.
Figure 4. Analysis of ubiquitinated protein levels in N2a cells in response to MG132 treatment.
Proteasome inhibitor treatment increases the levels of protein ubiquitination, and washout of the inhibitor sustains the elevation in ubiquitinated protein levels. Treatment with inhibitors of ERK1/2 and JNK have no significant effect on proteasome inhibitor-induced increases in ubiquitinated protein levels. Neural N2a cells were analyzed for the levels of ubiquitination following 6 hours of 10 μM MG132 treatment with or without the presence of 5 μM ERK1/2 inhibitor or 5 μM JNK inhibitor (AEG1842). Additionally, cultures were analyzed for N2a protein ubiquitination following washout of MG132 and incubation of cells in normal N2a media for 16 hours. Data representative of results from 3 separate experiments.
Effect of ERK1/2 and JNK inhibition on MG132-induced ribosome disturbances
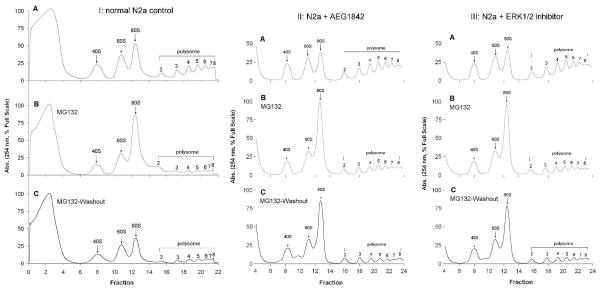
Studies from our laboratory and other have demonstrated that proteasome inhibition is sufficient to alter ribosome dysfunction and protein synthesis (Ding et al., 2006). In this study we observed that inhibition of proteasome activity induced declines in polyribosome levels and significantly altered the relative level of ribosome precursors (40S/60S) as well as mature ribosome complexes (80S) (Fig. 5). Washout of MG132 was observed to result in no significant effect on MG132 disturbances of either polyribosome and individual ribosome complexes (Fig. 5). Additionally, we observed that treatment with JNK inhibitor (Fig. 5) or ERK1/2 inhibitor (Fig. 5) did not have significant effects on MG132 induced ribosomal disturbances.
Figure 5. Analysis of MG132 effects on ribosome and polyribosome homeostasis in N2a neural cells.
Neural N2a cells were analyzed for alterations in ribosome and polysome fractions following 6 hour-treatment with 10 μM proteasome inhibitor MG132. Additionally, ribosome and polyribosome homeostasis was examined following washout of MG132 and incubation of cells in normal N2a media for 16 hours. Data demonstrate that MG132 results in diminution of polyribosome levels, and alters the ratio of premature (40S/60S) ribosome complexes to mature ribosomes (80S). Additional experiments were conducted where MG132 effects were analyzed in the presence and absence of 5 μM JNK inhibitor AEG1842. Lastly studies were conducted in which MG132 effects were analyzed in the presence and absence of 5 μM ERK1/2 inhibitor. Data representative of results from 3 separate experiments.
Discussion
Our findings indicate that N2a neural cells exhibit reversible proteasome inhibitor toxicity similar to what is observed in rat primary neurons (Ding et al., 2006), with washout of the reversible proteasome inhibitor largely ameliorating the neurotoxicity of proteasome inhibition. N2a cells may therefore provide another important and relevant model for studying the molecular and cellular aspects of proteasome inhibition, including identifying and understanding the reversible aspects of proteasome inhibitor toxicity. In particular, studies are needed to distinguish which downstream effects of proteasome inhibition are involved in promoting neurotoxicity, as well as identify those cellular disturbances which play a role in preventing neurotoxicity in response to proteasome inhibition. Such data are of significant importance in the neuroscience field given the considerable evidence for proteasome inhibition contributing to brain aging and neuropathology in a variety of neurodegenerative disorders (Keller et al., 2006; Olanow and McNaught, 2006; Stolzing and Grune, 2001; Sullivan et al., 2004; Halliwell, 2002; McNaught, 2004; Seo et al., 2004). Studies using N2a cells may therefore be combined with studies in primary rat neurons to develop useful insight into these areas of aging and neurodegeneration research.
The data in our current study demonstrate that proteasome inhibition promotes activation of ERK1/2 and JNK signal transduction pathways, and that the activation of these pathways appears to have differential effects on neural viability. Specifically, in the present study we identified that activation of ERK1/2 activation appears to play a significant role in ameliorating neurotoxicity while activation of JNK appears to promote the development of neural cell death. These studies are therefore consistent with previous studies which identified a role for ERK1/2 in ameliorating the toxicity of irreversible proteasome inhibitors (Choi et al., 2006; Cheng et al., 2006) and studies which identified JNK inhibitors as being capable of promoting cell viability in response to irreversible proteasome inhibitor administration (Shi et al., 2006; Cheng et al., 2006). At present it is unclear whether the ultimate fate of cell viability is dictated by the balance between the gross level of ERK1/2 activated events, and the gross level of JNK mediated events, or whether cell viability is dictated by the modulation of a limited number of specific events following activation of these signal transduction pathways. Interestingly, in the present study we observed that washout of the proteasome inhibitor resulted in a further increase in ERK1/2 activation while JNK activation was reduced in response to removal of proteasome inhibitor. These data suggest once proteasome inhibition has activated ERK1/2 and JNK pathways, ERK1/2 possesses significant residual activity and remains capable of sustaining downstream signal transduction, while maintenance of JNK activation requires sustained levels of proteasome inhibition. Understanding the basis for proteasome inhibition regulating these two kinases is therefore likely key to understanding the basis for proteasome inhibitor toxicity.
Previous studies have demonstrated that proteasome inhibition promotes an inhibtion of ERK1/2 activation via modulation of ERK1/2 specific phosphatases (Torres et al., 2003) in fibroblast. Furthermore, this previous study linked the ability of proteasome inhibitors to decrease ERK1/2 signaling to the ability of proteasome inhibitors to promote an acceleration in cell senescence (Torres et al., 2003), consistent with proteasome inhibition promoting cellular aging in some cell types via the down regulation of ERK 1/2 signaling. In our study we observed that neural cells have an apparent protective activation of ERK 1/2 in response to proteasome inhibition. It remains unclear as to why in some cell types there is a down regulation of ERK 1/2, and in others there is an apparent activation of ERK 1/2, in response to proteasome inhibition. The most obvious explanation based on the available data suggest that this discrepancy is largely dependent upon the degree to which MAPK phosphatase activity is increased in response to proteasome inhibition (Torres et al., 2003). Regardless, these studies cumulatively are consistent with ERK 1/2 activation being beneficial in ameliorating the effects of proteasome inhibition. Identifying the proteasome substrates which directly or indirectly are responsible for modulating ERK 1/2 activity, and the ERK 1/2 substrates which are responsible for modulating downstream effects of proteasome inhibition, will lead to significant advances in the area of aging research. Such studies may also explain the basis by which increasing the levels of proteasome in cells is sufficient to increase lifespan (Chen et al., 2006; Chondrogianni and Gonos, 2007, 2008; Chondrogianni et al., 2005).
It is important to point out that our studies relied on Western blot techniques to assess the relative activation state of ERK1/2 and JNK, with previous studies using these assays as well as other experimental modes to detect changes in ERK1/2 and JNK activity (Cheng et al., 2006). This discrepancy highlights the fact that there may be cell type specific, as well as experimental variances depending on the model utilized; with regards to the ultimate degree ERK1/2 and JNK are activated in response to proteasome inhibition. Such difference are important to consider when synthesizing the findings from different studies on this research topic, and may be important to understanding the ability of proteasome inhibition to promote cell death in some paradigms, and in other paradigms contribute to the induction of cell death.
Our data suggest that the toxicity of proteasome inhibition is regulated by the balance between pro- and anti-cell death downstream effects of proteasome inhibition. Previous studies have firmly established that proteasome inhibition results in increased levels of both pro- and anti-apoptotic pathways (Ding et al. 2004; Yew et al., 2005). This is an important concept for understanding how proteasome inhibition mediates neurotoxicity in vivo, given that it is likely that inhibition of proteasome activity in vivo is mediated by transient events that ultimately results in longer and longer durations of proteasome inhibition as tissues age. Identifying the neuropathological and neurochemical events which are central mediators of the deleterious shift to prolonged proteasome inhibition, will be useful in identifying cases where proteasome inhibition is a benign event.
Interestingly, in the present study we did not observe a significant role for JNK or ERK1/2 in modulating the levels of ubiquitinated protein or ribosome disturbances which occurred following proteasome inhibition. Such data suggest that these alterations may occur independent of increased ERK1/2 and JNK activity. Interestingly, these gross abnormalities were not clearly identified as being pro- or anti-cytotoxic given that increased protein ubiquitination and ribosome disturbances were observed during chronic as well as transient proteasome inhibitor treatment. These data suggest that these events are independent predictors of proteasome inhibitor toxicity, at least in this current experimental paradigm. Additionally, they raise the possibility that it may be small changes in specific pools of ubiquitinated protein, or specific effects in subsets of polyribosomes within a specific subcellular localization, which are linked to the induction of neurotoxicity. Such scenarios may be particularly important to the emerging role for proteasome activity in regulating synaptic homeostasis and long term potentiation (Ding and Shen, 2008; Fioravante et al., 2008). Our studies indicate that proteasome inhibition promotes the disassociation of polyribosome complexes, and thereby promotes the amount of free mature ribosomes in the cytoplasm. Such changes will undoubtedly contribute to impairments in protein synthesis, and are likely mediated by effects of proteasome inhibition on ribosomal RNA (Ding et al., 2006).
Given the tremendous interest in proteasome biology and interest in understanding the downstream effects of proteasome inhibition, it is becoming increasingly important to begin elucidating the basis by which dysfunction of the proteasome promotes cellular disturbances. In particular, studies need to begin to link the different downstream effects of proteasome inhibition to each other, and to begin the process of distinguishing benign effects from those which are either pro- and anti-apoptotic. Such data are likely to increase our understanding of the basis for brain aging and neurotoxicity in a variety of neurodegenerative settings where proteasome inhibition is observed.
Acknowledgments
This work was supported by grants from the National Institute of Aging (AG0257701, AG029885: J.N.K) and financial support from Hibernia National Bank/Edward G. Schlieder Endowed Chair (J.N.K). The authors have no conflicts of interest to report for the work presented in this manuscript.
References
- Borsello T, Forloni G. JNK signalling: a possible target to prevent neurodegeneration. Curr Pharm Des. 2007;13:1875–1886. doi: 10.2174/138161207780858384. [DOI] [PubMed] [Google Scholar]
- Breusing N, Grune T. Regulation of proteasome-mediated protein degradation during oxidative stress and aging. Biol Chem. 2008;389:203–209. doi: 10.1515/BC.2008.029. [DOI] [PubMed] [Google Scholar]
- Chen Q, Thorpe J, Dohmen JR, Li F, Keller JN. Ump1 extends yeast lifespan and enhances viability during oxidative stress: central role for the proteasome? Free Radic Biol Med. 2006;40:120–126. doi: 10.1016/j.freeradbiomed.2005.08.048. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Chang LW, Tsou TC. Mitogen-activated protein kinases mediate arsenic-induced down-regulation of survivin in human lung adenocarcinoma cells. Arch Toxicol. 2006;80:310–318. doi: 10.1007/s00204-005-0045-1. [DOI] [PubMed] [Google Scholar]
- Choi BH, Hur EM, Lee JH, Jun DJ, Kim KT. Protein kinase Cdelta-mediated proteasomal degradation of MAP kinase phosphatase-1 contributes to glutamate-induced neuronal cell death. J Cell Sci. 2006;119:1329–1340. doi: 10.1242/jcs.02837. [DOI] [PubMed] [Google Scholar]
- Chondrogianni N, Gonos ES. Proteasome dysfunction in mammalian aging: Steps and factors involved. Exp Gerontol. 2005;40:931–938. doi: 10.1016/j.exger.2005.09.004. [DOI] [PubMed] [Google Scholar]
- Chondrogianni N, Gonos ES. Overexpression of hUMP1/POMP proteasome accessory protein enhances proteasome-mediated antioxidant defence. Exp Gerontol. 2007;42:899–903. doi: 10.1016/j.exger.2007.01.012. [DOI] [PubMed] [Google Scholar]
- Chondrogianni N, Gonos ES. Proteasome activation as a novel antiaging strategy. IUBMB Life. 2008;60:651–655. doi: 10.1002/iub.99. [DOI] [PubMed] [Google Scholar]
- Chondrogianni N, Stratford FL, Trougakos IP, Friguet B, Rivett AJ, Gonos ES. Central role of the proteasome in senescence and survival of human fibroblasts: induction of a senescence-like phenotype upon its inhibition and resistance to stress upon its activation. J Biol Chem. 2003;278:28026–28037. doi: 10.1074/jbc.M301048200. [DOI] [PubMed] [Google Scholar]
- Chondrogianni N, Tzavelas C, Pemberton AJ, Nezis IP, Rivett AJ, Gonos ES. Overexpression of proteasome beta5 assembled subunit increases the amount of proteasome and confers ameliorated response to oxidative stress and higher survival rates. J Biol Chem. 2005;280:11840–11850. doi: 10.1074/jbc.M413007200. [DOI] [PubMed] [Google Scholar]
- de Vrij FM, Fischer DF, van Leeuwen FW, Hol EM. Protein quality control in Alzheimer’s disease by the ubiquitin proteasome system. Prog Neurobiol. 2004;74:249–270. doi: 10.1016/j.pneurobio.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Ding M, Shen K. The role of the ubiquitin proteasome system in synapse remodeling and neurodegenerative diseases. Bioessays. 2008;30:1075–1083. doi: 10.1002/bies.20843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Bruce-Keller AJ, Chen Q, Keller JN. Analysis of gene expression in neural cells subject to chronic proteasome inhibition. Free Radic Biol Med. 2004;36:445–455. doi: 10.1016/j.freeradbiomed.2003.10.025. [DOI] [PubMed] [Google Scholar]
- Ding Q, Cecarini V, Keller JN. Interplay between protein synthesis and degradation in the CNS: physiological and pathological implications. Trends Neurosci. 2007;30:31–36. doi: 10.1016/j.tins.2006.11.003. [DOI] [PubMed] [Google Scholar]
- Ding Q, Dimayuga E, Markesbery WR, Keller JN. Proteasome inhibition induces reversible impairments in protein synthesis. FASEB J. 2006;20:1055–1063. doi: 10.1096/fj.05-5495com. [DOI] [PubMed] [Google Scholar]
- Ding Q, Dimayuga E, Keller JN. Proteasome regulation of oxidative stress in aging and age-related diseases of the CNS. Antioxid Redox Signal. 2006;8:163–172. doi: 10.1089/ars.2006.8.163. [DOI] [PubMed] [Google Scholar]
- Fineschi S, Bongiovanni M, Donati Y, Djaafar S, Naso F, Goffin L, Argiroffo CB, Pache JC, Dayer JM, Ferrari-Lacraz S, Chizzolini C. In vivo investigations on anti-fibrotic potential of proteasome inhibition in lung and skin fibrosis. Am J Respir Cell Mol Biol. 2008;39:458–465. doi: 10.1165/rcmb.2007-0320OC. [DOI] [PubMed] [Google Scholar]
- Fioravante D, Liu RY, Byrne JH. The ubiquitin-proteasome system is necessary for long-term synaptic depression in Aplysia. J Neurosci. 2008;28:10245–10256. doi: 10.1523/JNEUROSCI.2139-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AL. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426:895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- Gray DA, Tsirigotis M, Woulfe J. Ubiquitin, proteasomes, and the aging brain. Sci Aging Knowledge Environ 2003. 2003:RE6. doi: 10.1126/sageke.2003.34.re6. [DOI] [PubMed] [Google Scholar]
- Grune T, Jung T, Merker K, Davies KJ. Decreased proteolysis caused by protein aggregates, inclusion bodies, plaques, lipofuscin, ceroid, and ‘aggresomes’ during oxidative stress, aging, and disease. Int J Biochem Cell Biol. 2004;36:2519–2530. doi: 10.1016/j.biocel.2004.04.020. [DOI] [PubMed] [Google Scholar]
- Hyun DH, Lee M, Halliwell B, Jenner P. Proteasomal inhibition causes the formation of protein aggregates containing a wide range of proteins, including nitrated proteins. J Neurochem. 2003;86:363–367. doi: 10.1046/j.1471-4159.2003.01841.x. [DOI] [PubMed] [Google Scholar]
- Jiang HY, Wek RC. Phosphorylation of the alpha-subunit of the eukaryotic initiation factor-2 (eIF2alpha) reduces protein synthesis and enhances apoptosis in response to proteasome inhibition. J Biol Chem. 2005;280:14189–14202. doi: 10.1074/jbc.M413660200. [DOI] [PubMed] [Google Scholar]
- Junttila MR, Li SP, Westermarck J. Phosphatase-mediated crosstalk between MAPK signaling pathways in the regulation of cell survival. FASEB J. 2008;22:954–965. doi: 10.1096/fj.06-7859rev. [DOI] [PubMed] [Google Scholar]
- Keller JN, Hanni KB, Markesbery WR. Impaired proteasome function in Alzheimer’s disease. J Neurochem. 2000;75:436–439. doi: 10.1046/j.1471-4159.2000.0750436.x. [DOI] [PubMed] [Google Scholar]
- Keller JN, Gee J, Ding Q. The proteasome in brain aging. Ageing Res Rev. 2002;1:279–293. doi: 10.1016/s1568-1637(01)00006-x. [DOI] [PubMed] [Google Scholar]
- Keller JN, Kindy MS, Holtsberg FW, St Clair DK, Yen HC, Germeyer A, Steiner SM, Bruce-Keller AJ, Hutchins JB, Mattson MP. Mitochondrial manganese superoxide dismutase prevents neural apoptosis and reduces ischemic brain injury: suppression of peroxynitrite production, lipid peroxidation, and mitochondrial dysfunction. J Neurosci. 1998;18:687–697. doi: 10.1523/JNEUROSCI.18-02-00687.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TS, Jang CY, Kim HD, Lee JY, Ahn BY, Kim J. Interaction of Hsp90 to ribosomal proteins protects from ubiquitination and proteasome-dependent degradation. Mol Biol Cell. 2005;17:824–833. doi: 10.1091/mbc.E05-08-0713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruman I, Bruce AJ, Bredesen DE, Waeg G, Mattson MP. Evidence that 4-hydroxynonenal mediates oxidative stress-induced neuronal apoptosis. J Neurosci. 1997;17:5089–5100. doi: 10.1523/JNEUROSCI.17-13-05089.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell B. Hypothesis: proteasomal dysfunction a primary event in neurodegeneration that leads to nitrative and oxidative stress and subsequent neuron death. Ann N Y Acad Sci. 2002;962:182–194. doi: 10.1111/j.1749-6632.2002.tb04067.x. [DOI] [PubMed] [Google Scholar]
- Harris GF, 4th, Anderson ME, Lee JH. The effect of proteasome inhibition on p53 degradation and proliferation in tonsil epithelial cells. Arch Otolaryngol Head Neck Surg. 2008;134:157–163. doi: 10.1001/archoto.2007.37. [DOI] [PubMed] [Google Scholar]
- Lam PY, Cadenas E. Compromised proteasome degradation elevates neuronal nitric oxide synthase levels and induces apoptotic cell death. Arch Biochem Biophys. 2008;478:181–186. doi: 10.1016/j.abb.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauricella M, Emanuele S, D’Anneo A, Calvaruso G, Vassallo B, Carlisi D, Portanova P, Vento R, Tesoriere G. JNK and AP-1 mediate apoptosis induced by bortezomib in HepG2 cells via FasL/caspase-8 and mitochondria-dependent pathways. Apoptosis. 2006;11:607–625. doi: 10.1007/s10495-006-4689-y. [DOI] [PubMed] [Google Scholar]
- Li X, Du Y, Fan X, Yang D, Luo G, Le W. c-Jun N-terminal kinase mediates lactacystin-induced dopamine neuron degeneration. J Neuropathol Exp Neurol. 2008;67:933–944. doi: 10.1097/NEN.0b013e318186de64. [DOI] [PubMed] [Google Scholar]
- Li F, Zhang L, Craddock J, Bruce-Keller AJ, Dasuri K, Nguyen A, Keller JN. Aging and dietary restriction effects on ubiquitination, sumoylation, and the proteasome in the heart. Mech Ageing Dev. 2008;129:515–521. doi: 10.1016/j.mad.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam PY, Cadenas E. Compromised proteasome degradation elevates neuronal nitric oxide synthase levels and induces apoptotic cell death. Arch Biochem Biophys. 2008;478:181–186. doi: 10.1016/j.abb.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YH, Wei W, Yin J, Liu GP, Wang Q, Cao FY, Wang JZ. Proteasome inhibition increases tau accumulation independent of phosphorylation. Neurobiol Aging. 2008 doi: 10.1016/j.neurobiolaging.2008.02.012. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- McNaught KS. Proteolytic dysfunction in neurodegenerative disorders. Int Rev Neurobiol. 2004;62:95–119. doi: 10.1016/S0074-7742(04)62003-4. [DOI] [PubMed] [Google Scholar]
- Meiners S, Ludwig A, Stangl V, Stangl K. Proteasome inhibitors: poisons and remedies. Med Res Rev. 2008;28:309–327. doi: 10.1002/med.20111. [DOI] [PubMed] [Google Scholar]
- Montagut C, Rovira A, Albanell J. The proteasome: a novel target for anticancer therapy. Clin Transl Oncol. 2006;28:313–317. doi: 10.1007/s12094-006-0176-8. [DOI] [PubMed] [Google Scholar]
- Olanow CW, McNaught KS. Ubiquitin-proteasome system and Parkinson’s disease. Mov Disord. 2006;21:1806–1823. doi: 10.1002/mds.21013. [DOI] [PubMed] [Google Scholar]
- Othumpangat S, Kashon M, Joseph P. Eukaryotic translation initiation factor 4E is a cellular target for toxicity and death due to exposure to cadmium chloride. J Biol Chem. 2005;280:25162–25169. doi: 10.1074/jbc.M414303200. [DOI] [PubMed] [Google Scholar]
- Raman M, Chen W, Cobb MH. Differential regulation and properties of MAPKs. Oncogene. 2007;26:3100–3112. doi: 10.1038/sj.onc.1210392. [DOI] [PubMed] [Google Scholar]
- Rideout HJ, Larsen KE, Sulzer D, Stefanis L. Proteasomal inhibition leads to formation of ubiquitin/alpha-synuclein-immunoreactive inclusions in PC12 cells. J Neurochem. 2001;78:899–908. doi: 10.1046/j.1471-4159.2001.00474.x. [DOI] [PubMed] [Google Scholar]
- Rideout HJ, Wang Q, Park DS, Stefanis L. Cyclin-dependent kinase activity is required for apoptotic death but not inclusion formation in cortical neurons after proteasomal inhibition. J Neurosci. 2003;23:1237–1245. doi: 10.1523/JNEUROSCI.23-04-01237.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo H, Sonntag KC, Isacson O. Generalized brain and skin proteasome inhibition in Huntington’s disease. Ann Neurol. 2004;56:319–328. doi: 10.1002/ana.20207. [DOI] [PubMed] [Google Scholar]
- Shah IM, Di Napoli M. The ubiquitin-proteasome system and proteasome inhibitors in central nervous system diseases. Cardiovasc Hematol Disord Drug Targets. 2007;7:250–273. doi: 10.2174/187152907782793572. [DOI] [PubMed] [Google Scholar]
- Shi YY, Small GW, Orlowski RZ. Proteasome inhibitors induce a p38 mitogen-activated protein kinase (MAPK)-dependent anti-apoptotic program involving MAPK phosphatase-1 and Akt in models of breast cancer. Breast Cancer Res Treat. 2006;100:33–47. doi: 10.1007/s10549-006-9232-x. [DOI] [PubMed] [Google Scholar]
- Shringarpure R, Davies KJ. Protein turnover by the proteasome in aging and disease. Free Radic Biol Med. 2002;32:84–88. doi: 10.1016/s0891-5849(02)00824-9. [DOI] [PubMed] [Google Scholar]
- Stolzing A, Grune T. The proteasome and its function in the ageing process. Clin Exp Dermatol. 2001;26:566–572. doi: 10.1046/j.1365-2230.2001.00867.x. [DOI] [PubMed] [Google Scholar]
- Sullivan PG, Dragicevic NB, Deng JH, Bai Y, Dimayuga E, Ding Q, Chen Q, Bruce-Keller AJ, Keller JN. Proteasome inhibition alters neural mitochondrial homeostasis and mitochondria turnover. J Biol Chem. 2004;279:20699–20707. doi: 10.1074/jbc.M313579200. [DOI] [PubMed] [Google Scholar]
- Sun F, Kanthasamy A, Song C, Yang Y, Anantharam V, Kanthasamy AG. Proteasome inhibitor-induced apoptosis is mediated by positive feedback amplification of PKCdelta proteolytic activation and mitochondrial translocation. J Cell Mol Med. 2008;12:2467–2481. doi: 10.1111/j.1582-4934.2008.00293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SJ, Loftus LT, Ashley MD, Meller R. Ubiquitin-proteasome system as a modulator of cell fate. Curr Opin Pharmacol. 2008;8:90–85. doi: 10.1016/j.coph.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres C, Francis MK, Lorenzini A, Tresini M, Cristofalo VJ. Metabolic stabilization of MAP kinase phosphatase-2 in senescence of human fibroblasts. Exp Cell Res. 2003;290:195–206. doi: 10.1016/s0014-4827(03)00309-4. [DOI] [PubMed] [Google Scholar]
- Torres C, Lewis L, Cristofalo VJ. Proteasome inhibitors shorten replicative life span and induce a senescent-like phenotype of human fibroblasts. J Cell Physiol. 2006;207:845–853. doi: 10.1002/jcp.20630. [DOI] [PubMed] [Google Scholar]
- Torres CA, Perez VI. Proteasome modulates mitochondrial function during cellular senescence. Free Radic Biol Med. 2008;44:403–414. doi: 10.1016/j.freeradbiomed.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu HY, Juvekar A, Ghosh C, Ramaswami S, Le DH, Vancurova I. Proteasome inhibitors induce apoptosis of prostate cancer cells by inducing nuclear translocation of IkappaBalpha. Arch Biochem Biophys. 2008;475:156–163. doi: 10.1016/j.abb.2008.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Hoshino Y, Ito T, Nariai T, Mohri T, Obana M, Hayata N, Uozumi Y, Maeda M, Fujio Y, Azuma J. Atrogin-1 ubiquitin ligase is upregulated by doxorubicin via p38-MAP kinase in cardiac myocytes. Cardiovasc Res. 2008;279:89–96. doi: 10.1093/cvr/cvn076. [DOI] [PubMed] [Google Scholar]
- Yew EH, Cheung NS, Choy MS, Qi RZ, Lee AY, Peng ZF, Melendez AJ, Manikandan J, Koay ES, Chiu LL, Ng WL, Whiteman M, Kandiah J, Halliwell B. Proteasome inhibition by lactacystin in primary neuronal cells induces both potentially neuroprotective and pro-apoptotic transcriptional responses: a microarray analysis. J Neurochem. 2005;94:943–956. doi: 10.1111/j.1471-4159.2005.03220.x. [DOI] [PubMed] [Google Scholar]
- Zhou D, Palam LR, Jiang L, Narasimhan J, Staschke KA, Wek RC. Phosphorylation of eIF2 Directs ATF5 Translational Control in Response to Diverse Stress Conditions. J Biol Chem. 2008;283:7064–7073. doi: 10.1074/jbc.M708530200. [DOI] [PubMed] [Google Scholar]