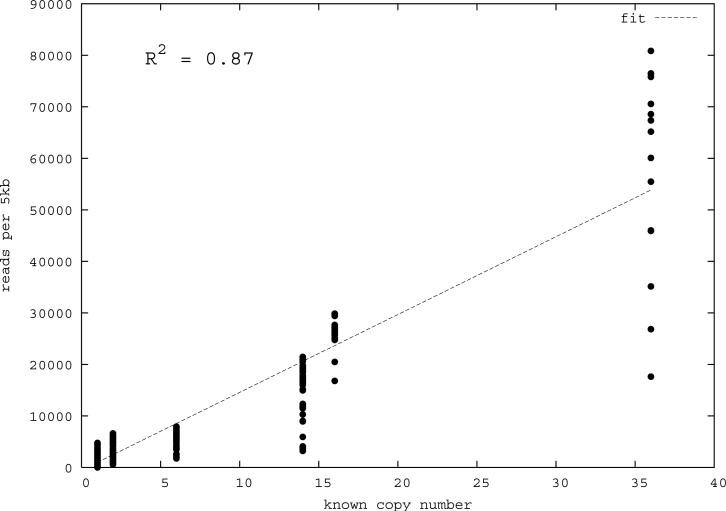
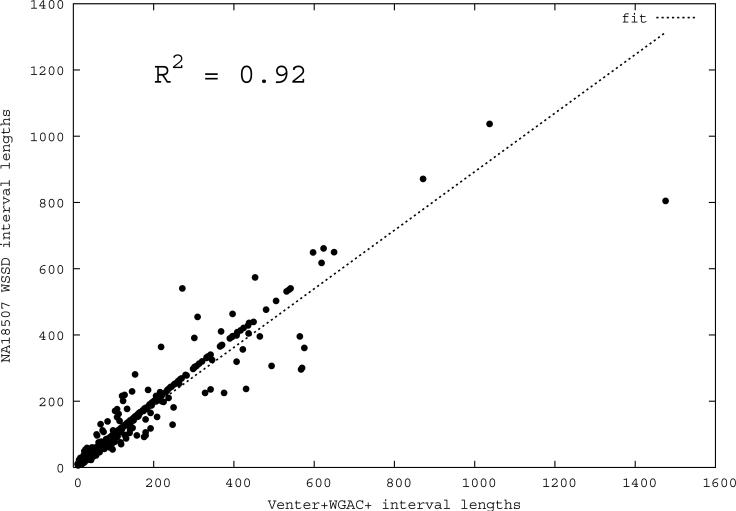
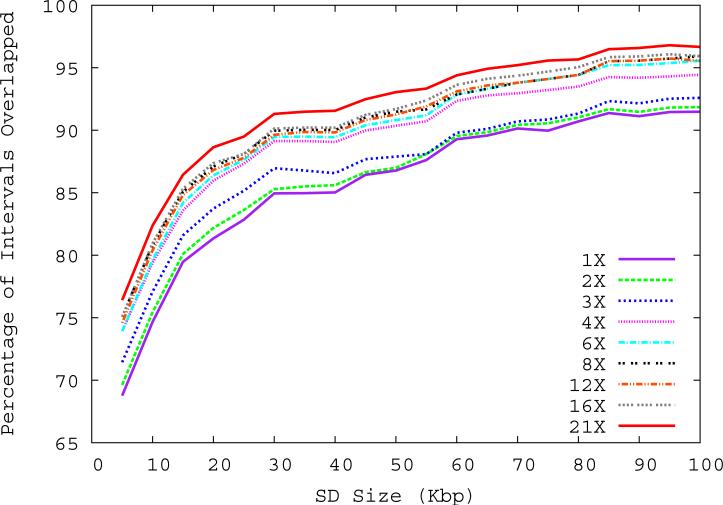
Figure 1. Correlation of predicted and known segmental duplications (NA18507).
a) mrFAST sequence read-depth per 5-kbp window along the human genome correlates well (R2=0.87) with the known copy number of duplicated sequences. b) Predicted duplication interval length versus the assembly-based length intervals of known duplications (Whole Genome Assembly Comparison; WGAC, ≥94% sequence identity) 34 shows that boundaries of duplications can be accurately predicted. A few intervals show discrepancy in boundary prediction, however, this is largely due to deletion polymorphism in the NA18507 genome within duplications (supported by arrayCGH). c) A cumulative plot of the fraction of duplication intervals detected as a function of various read-depth sequence coverage. The segmental duplication (SD) size is given in cumulative intervals (≥5 kbp, ≥10 kbp, etc.) and represents the set of intervals identified both within the public reference assembly (build35) and the Celera whole-genome shotgun sequence reads. As expected, the sensitivity of our method increases with more genome coverage; the most dramatic difference in detection is observed between 3- to 4-fold coverage.