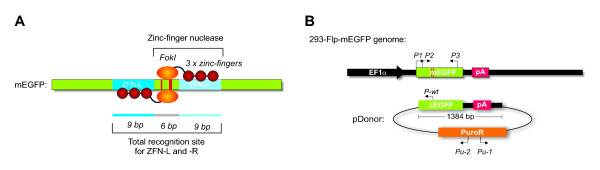
Figure 1.
Schematic representation of the model system. (A) Features of the mEGFP coding sequence (green) with binding sites for ZFN-Left (L) and ZFN-Right (R) indicated (dark and light blue, respectively). The positions of the mutations rendering the mEGFP gene non-functional are shown in red. The ZFN-L and ZFN-R bound to their target sequences are indicated, each with their zinc-finger domains (3 × red circles) and the FokI nuclease domain (orange circle). The two 9 bp long recognition sites for ZFN-L (GGG GTA CGC) and ZFN-R (GAA GCA GCA) are separated by a 6 bp spacer region required for optimal positioning of the two FokI nuclease domains. Given that the FokI nuclease domain must dimerize in order for a DNA DSB to be formed, the total recognition sequence required for DSB formation is 18 bp, excluding the 6 bp spacer region. The 293-Flp-mEGFP cell line contained a single copy chromosomally integrated mEGFP gene. (B) Arrangement of the mEGFP gene and surrounding genetic elements in the 293-Flp-mEGFP cell line including the human Elongation Factor 1a promoter (EF1a, black), the mEGFP gene (green) with the point mutations (red) and the poly-adenylation signal (pA, red). The pDonor construct, provided as a substrate for HR, is shown as a circle with the regions homologous to the 293-Flp-mEGFP genomic sequence as indicated (ΔEGFP and pA, 1384 bp in total). The expression cassette for the PuroR gene is outlined in orange. Position and direction of the PCR primers used (P1, P2, P3, P-wt, Pu-1 and Pu-2) are indicated. With this model system, HR between the genomic mEGFP gene in the 293-Flp-mEGFP cells and the ΔEGFP gene in the pDonor construct could be measured by quantification of green fluorescent cells, while the rate of illegitimate integration could be quantified by appearance of puromycin resistant clones.