Abstract
Comparative genomic hybridization studies have revealed elevated copy number (CN) at the reticulocyte binding protein 1 gene (PfRh1) in fast growing lab-adapted parasites, while genetic manipulation demonstrates a causal link between cell invasion and PfRh1 CN. We therefore examined PfRh1 copy number variation (CNV) in 202 single clone parasite isolates from four countries to quantify the extent of CNV within natural populations. Surprisingly, we found that no natural parasite infections showed elevated CN. In contrast, 4/28 independent laboratory reference strains show elevated CN. One possibility is that amplification of PfRh1 (or neighboring loci) is selected during laboratory culture. In the case of FCR3-group of parasites, clone trees show that PfRh1 amplification arose in laboratory lines following establishment in culture. These data show that CNV at PfRh1 is rare or non-existent in natural populations, but can arise during laboratory propagation. We conclude that PfRh1 CNV is not an important determinant of gene expression, cell invasion or growth rate in natural parasite populations.
Domesticated animals differ in many ways from their wild ancestors [1;2]. In the same way laboratory adaptation of malaria parasites (Plasmodium falciparum) may result in genotypic and phenotypic changes, as parasites are selected for rapid growth, and freed from immune attack and the constraints of sexual reproduction. For example, subtelomeric deletions on chr 2 and 9 occur repeatedly during laboratory adaptation and result in loss of cytoadherence [3;4]. As a consequence care is required when making conclusions about malaria biology based on laboratory-adapted isolates. We provide a cautionary tale relating to another important phenotype – red blood cell invasion - to illustrate this.
Parasite growth rate is an important determinant of malaria pathogenesis and varies considerably between parasite lines [5]. Merozoite invasion is orchestrated by two protein families, the Duffy binding-like (DBL) family and the reticulocyte-binding proteins (RBL) [6;7]. The RBL protein PfRh1 is required for sialic acid dependent (neuramidase-sensitive) invasion of human erythrocytes. All parasite strains express this protein, but parasite lines differ in their level of expression [7-9]. Laboratory strains such as FCR3 and FCB show elevated gene expression which is as a consequence of amplification of PfRh1 gene CN [7]. Gene disruption experiments show that reduction in PfRh1 CN does not block invasion but cause parasites to invade cells using alternative neuramidase-resistant pathways, demonstrating a causal relationship between PfRh1 gene dosage and cell invasion [7]. Comparative genomic hybridization studies further support the idea that PfRh1 CN may influence growth rate. Ribacke et al [10] examined 12 parasite isolates. They found 5-7 fold amplification of PfRh1 in two rapidly growing parasites (FCR3 and F32), and suggested that PfRh1 amplification might underlie rapid growth.
We are interested in variation in gene CN, particularly when it generates changes in phenotype. We therefore decided to examine CNV at PfRh1 in both field collected and laboratory isolates. We collected blood samples from patients in SE Asia (Thailand and Lao PDR (Laos)) and Africa (Uganda and Malawi). Thai samples were collected at the Mawker-Thai clinic on the Thai-Burma border (December 2000 - September 2003). We collected samples from Phalanxay (Savannaket Province, Laos) between June 2002 and Sept 2003 from patients involved in clinical drug efficacy trials. P. falciparum isolates were collected from children under five years of age presenting to the Queen Elizabeth Central Hospital, Blantyre, Malawi with uncomplicated falciparum malaria in 2008. In Uganda, P. falciparum –infected blood samples were collected (April 1996–June 1997) from HIV-infected individuals living in a 15-km radius of a clinic near Entebbe. In all cases, parasite DNA was prepared by phenol/chloroform extraction of whole blood, following removal of buffy coats. Collection protocols were approved by regional ethics committees and by the Institutional Review Board at the University of Texas Health Science Center at San Antonio.
Initially we genotyped all isolates using seven microsatellite loci to exclude multiple clone infections, leaving 202 parasite isolates with a single clone [11]. These included 107 from Thailand, 42 from Laos, 19 from Malawi and 34 from Uganda. We also obtained DNA from 49 laboratory isolates from the Malaria Research and Reference Reagent Resource Center (MR4) (Manassas, VA, USA) (Supplementary Table S1). These were also genotyped using the seven microsatellites. We found many identical multilocus genotypes, consistent with analyses using single nucleotide polymorphisms (SNPs) [12]. Of 49 parasite lines, there were 28 independent genotypes. For example, two pairs of parasites WR87 and MT/S1, and TM90C6A and TM93C1088 were indistinguishable. Similarly a group of 11 isolates comprising FCR3 and derived clones, FCB, ITG and T9/94 had the same multilocus genotype consistent with a common origin (Supplementary Table S1), most likely from a SE Asian source [13]. We focus on this group of isolates (the FCR3 group) that have clearly derived from a common ancestor, but have been maintained in a number of different laboratories under different names and selection regimes.
We used a real-time PCR assay to measure CN of PfRh1 (PFD0110w) relative to a single copy gene (PFL0770w: Seryl-T synthetase). We used the ΔΔCT method to measure CN relative to a calibrator sample (3D7). Assay details are provided in Table 1. The assay was experimentally validated as the efficiencies of the target and reference were approximately equal. To validate our real-time PCR assay we compared a parasite (FCR3CSA) known to have multiple copies of RH1 [7] with parasites thought to have a single copy (3D7). The real time assay estimated 4.59 copies (95% CI 4.09-5.15) in FCR3CSA (MRA-321G) relative to 3D7.
Table 1. Real time PCR assay for PfRh1.
We ran single plex assays on a Applied Biosystems 7900HT real-time PCR machine using standard conditions and Minor groove binding (MGB) probes. Both test and endogenous control gene PCRs were conducted in quadruplicate in 10μl reactions. We measured relative CN using the ΔΔCT method, which compares ΔCT (the difference in CT values between PfRh1 and control genes) between test DNA samples and a calibrator sample with known CN (ABI User Bulletin 2 http://www3.appliedbiosystems.com/cms/groups/mcb_support/documents/generaldocuments/cms_040980.pdf). The calibrator used was 3D7 which has PfRh1 CN of 1. CN of the test sample is estimated as 2 −ΔΔCT, while the 95% confidence intervals around CN estimates were derived from the variance among the four replicate reactions. As PfRh1 gene is highly polymorphic, we designed primers and probe within a region (chr 4: 146134-146968) that is devoid of SNPs in sequences available on Plasmodb (www.plasmdb.org). We excluded realtime PCR data when the CT was > 32 for either test or reference gene, or if the upper 95% confidence interval/ CN estimate > 0.4, or if the estimated CN was <0.5 as described previously [11].
| Gene | Primers | Probe sequence |
|---|---|---|
| Test gene: | CCAAAATCTTCTTAAATCCTTGTATA | 6FAM-ACACATTCATAGACACAATT |
| PfRh1 (PFD0110w) | TTTTTTCGTCTGTATAAACGTGTTGTG | |
| Endogenous control: | TTAGATTTTCAAGCGAGACGTTTAAA | VIC-CCAATAATTTCTGCCATACTA |
| Seryl-T synthetase (PFL0770w) | CCTTCCTACGGCTAAACCTGAAC |
To further demonstrate that the real-time assay results were not influenced by sequence variation, we used custom cGH microarrays (RocheNimblegen) printed with 385,585 probes of mean length 56 bp, spaced on average at 21bp intervals across the P. falciparum (3D7) genome [14]. Labeling and hybridization were done following standard protocols and data were analyzed using NIMBLESCAN and SIGNALMAP software. These analyses demonstrate that the amplified region in FCR3/Gambia Clone D4 spans 31.5kb on chr 4 and contains four genes, while the W2 amplicon spans 74.6 kb and contains 14 genes, consistent with previous observations [10;15]. The CN estimates from microarrays correspond closely with those from real-time PCR. Log2 ratios provide CN estimates of 2 for W2 and 5.3 for FCR3/Gambia Clone D4, consistent with real-time estimates of 2 and 7 respectively. Similarly, Dd2 and FCR-3/FMG (Gambia) are clearly single copy, consistent with real-time PCR estimates. The close correspondence between CN estimates from our real-time PCR assay and microarray data provides strong validation for our real-time PCR assay.
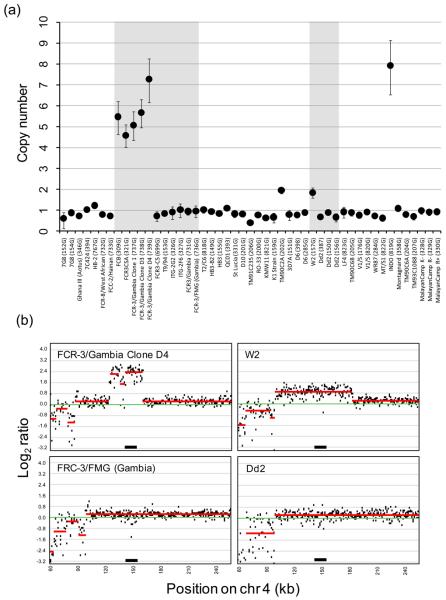
We used the validated real-time assay to measure PfRh1 CNV in the 202 field isolates. Surprisingly, none of these showed multiple copies of PfRh1. These data suggest that PfRh1 CNV is not an important determinant of invasion pathway or growth rate in the populations examined and is consistent with surveys of field isolates from Kenyan [8] and Senegal [16]. The extensive variation in expression of PfRh1 in parasites from patients [8;9] is due to factors other than gene-dosage. Our sampling does not include P. falciparum from South America, Papua New Guinea, or India, so we cannot exclude the possibility that PfRh1 amplification occurs in these locations. In contrast, 8/49 (16%) laboratory isolates, or 4/28 (14%) independent genotypes showed multiple copies (Figure 1, Table S1). These parasites carried an estimated 1.84 copies in W2 (MRA-157G) to 7.9 copies in INDO (MRA-819G). The difference in frequency of samples showing amplified PfRh1 between field collected and laboratory samples is highly significant (2-tailed Fishers exact test: p=0.0002), suggesting that amplification may be selected during laboratory culture (Fig 1b). The target of selection may be PfRh1 or a neighboring gene.
Figure 1. Copy number amplification at PfRh1.
(a) Real-time CN estimates for laboratory strains. The error bars show 95% confidence intervals around estimates of CN. The grey boxes shows strains in the FCR3 or W2/Dd2 group that are genetically identical for microsatellites (this study) and SNPs [12], but differ in PfRh1 CN. Details of the parasites lines used and microsatellite genotypes are shown in Supplementary Table S1. (b) Microarray validation of real-time PCR. Log2 plots of chr 4 for closely related parasites with and without amplification at PfRh1. Points show average Log2 ratios of probes over 500bp windows, while red lines show CN estimates of genome blocks. The two left hand panels show plots for FCR-3/Gambia Clone D4 (7.2 copies by real-time PCR) and FRC-3/FMG (Gambia) (1 copy); the right hand panels show W2 (two copies) and Dd2 (one copy). The amplified region of FCR-3/Gambia Clone D4 and W2 contain 4 and 14 genes respectively and are consistent with a previous report [15]. The position of PfRh1 is shown by a black bar.
The FCR3 group of parasites is of particular interest and provides evidence that amplification has occurred during laboratory culture. Five of eleven parasites in this group (FCB, FCR3CSA, FCR-3/Gambia Clone 1, FCR-3/Gambia Clone D3, FCR-3/Gambia Clone D-4,Knobless) show amplification (CN = 4.6-7.3), while the remaining six (FCR3-C5, T9/94, ITG-2G2, ITG-2F6, FCR3/Gambia, FCR-3/FMG (Gambia)) have a single copy (Fig 2). The dimorphism in CN within this group suggests either that PfRh1 amplification was present in the common ancestor and has subsequently been lost, or that the common ancestor had a single copy and amplification occurred during laboratory propagation. Reconstruction of a clone tree strongly supports that amplification occurred during parasite culture (Figure 2). A strain of P. falciparum (FMG) obtained in 1976 from Fajara, Gambia was cultured continuously giving rise to the line FCR3/FMG. The parasite cultured in the same lab also came to be known as FCR3/Gambia [17]. Both parasite lines have a single copy of the PfRh1 gene. FCR3/Gambia was maintained in culture for five years. In 1981 they were selected and cloned giving rise to FCR-3/Gambia Clone 1, FCR-3 Clone D3, and FCR-3 Clone D4 [18]. These strains have multiple copies of PfRh1. By implication amplification of PfRh1 occurred prior to selection of sub clones between 1976 and 1981.
Figure 2. Clone tree for 5 isolates in the FCR3 group.
The asterisk shows where we hypothesize that the amplification of PfRh1 occurred. The other members of the FCR3 group cannot be placed with certainty. The fact that parasites in this group were initially established from a Gambian source, but are clearly placed by SNP analysis within a clade of SE Asian samples [13;20] strongly suggests a contaminant origin for all parasites in this group.
It is important to note that the region of chr 4 amplified in the FCR3 group parasites contains three other genes in addition to PfRh1. These are surface-associated interspersed gene (SURFIN4.1, PFD0100c), and two exported protein of unknown function (PFD0095c and PFD0115c). It is conceivable that selection for increased dosage one of one of these genes drives amplification rather that PfRh1 itself.
The W2/Dd2 group of parasites also suggests change in PfRh1 CN has occurred during laboratory propagation. Dd2 is a mefloquine resistant clone derived from W2mef, a parasite line derived from W2 [19]. While three different Dd2 stocks from MR4 have a single copy, W2 clearly has 2 copies. In this case it is unclear whether CN was reduced in Dd2 after its derivation from W2, or if W2 has gained copies following the split with Dd2.
Elevated CN of chromosome regions containing PfRh1 is found in only 4/28 independent lab isolates. If this amplification is adaptive in laboratory culture, why has it not evolved in more parasite lines? We suggest three possible explanations. First, fitness effects of amplification may be dependent on genetic background, and may be deleterious in some parasite isolates. Second, duplication rates may differ between parasite lines, perhaps due to the length and position of nearly repeat sequences, as these act as templates for chromosome misalignment during DNA replication [4]. Third, fitness effects of duplication may be dependent on the cell invasion pathway used. These explanations are speculative, but all are experimentally testable.
In conclusion, while we do not find PfRh1 amplification in natural populations, we found that a higher proportion of laboratory isolates showed PfRh1 amplification, consistent with changes in CN during laboratory propagation. Strong support for this notion comes from observed dimorphism of PfRh1 CN in the FCR3 group of parasites. We conclude that gene dosage at this locus does not play an important role in explaining natural variation in expression, invasion and growth rates. Furthermore, we caution that surveys of CNV should avoid samples maintained in laboratory culture, and that biological insights gained from laboratory isolates should be verified where possible in field isolates.
Supplementary Material
Acknowledgements
Supported by NIH R01 AI075145 and R01 AI48071 (TJCA). The molecular work at the Southwest Foundation for Biomedical Research was conducted in facilities constructed with support from Research Facilities Improvement Program Grant C06 RR013556 from the National Center for Research Resources, National Institutes of Health. Collection of samples in Laos was funded by the Wellcome Trust of Great Britain. We thank Paul Newton for assistance in obtaining samples and the Ferdig laboratory (Notre Dame) for training in microarray methods, and for allowing us to use their array design. Suggestions from Ian Cheeseman improved the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Goodrich J, Wiener P. A walk from the wild side: the genetics of domestication of livestock and crops. Bioessays. 2005;27:574–6. doi: 10.1002/bies.20228. [DOI] [PubMed] [Google Scholar]
- 2.Wayne RK, Ostrander EA. Lessons learned from the dog genome. Trends Genet. 2007;23:557–67. doi: 10.1016/j.tig.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 3.Biggs BA, Kemp DJ, Brown GV. Subtelomeric chromosome deletions in field isolates of Plasmodium falciparum and their relationship to loss of cytoadherence in vitro. Proc.Natl.Acad.Sci U.S.A. 1989;86:2428–32. doi: 10.1073/pnas.86.7.2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson TJ, Patel J, Ferdig MT. Gene copy number and malaria biology. Trends Parasitol. 2009;25:336–43. doi: 10.1016/j.pt.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reilly HB, Wang H, Steuter JA, Marx AM, Ferdig MT. Quantitative dissection of clone-specific growth rates in cultured malaria parasites. Int.J.Parasitol. 2007;37:1599–607. doi: 10.1016/j.ijpara.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cowman AF, Crabb BS. Invasion of red blood cells by malaria parasites. Cell. 2006;124:755–66. doi: 10.1016/j.cell.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 7.Triglia T, Duraisingh MT, Good RT, Cowman AF. Reticulocyte-binding protein homologue 1 is required for sialic acid-dependent invasion into human erythrocytes by Plasmodium falciparum. Mol.Microbiol. 2005;55:162–74. doi: 10.1111/j.1365-2958.2004.04388.x. [DOI] [PubMed] [Google Scholar]
- 8.Nery S, Deans AM, Mosobo M, Marsh K, Rowe JA, Conway DJ. Expression of Plasmodium falciparum genes involved in erythrocyte invasion varies among isolates cultured directly from patients. Mol Biochem.Parasitol. 2006;149:208–15. doi: 10.1016/j.molbiopara.2006.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gomez-Escobar N, Amambua-Ngwa A, Walther M, Okebe J, Ebonyi A, Conway DJ. Erythrocyte invasion and merozoite ligand gene expression in severe and mild Plasmodium falciparum malaria. J.Infect.Dis. 2010;201:444–52. doi: 10.1086/649902. [DOI] [PubMed] [Google Scholar]
- 10.Ribacke U, Mok BW, Wirta V, et al. Genome wide gene amplifications and deletions in Plasmodium falciparum. Mol.Biochem.Parasitol. 2007;155:33–44. doi: 10.1016/j.molbiopara.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 11.Anderson TJ, Nair S, Qiu WG, et al. Are transporter genes other than the chloroquine resistance locus (pfcrt) and multi drug resistance gene (pfmdr) associated with antimalarial drug resistance? Antimicrob.Agents Chemother. 2005;272:1153–61. doi: 10.1128/AAC.49.6.2180-2188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daniels R, Volkman SK, Milner DA, et al. A general SNP-based molecular barcode for Plasmodium falciparum identification and tracking. Malar.J. 2008;7:223. doi: 10.1186/1475-2875-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neafsey DE, Schaffner SF, Volkman SK, et al. Genome-wide SNP genotyping highlights the role of natural selection in Plasmodium falciparum population divergence. Genome Biol. 2008;9:R171. doi: 10.1186/gb-2008-9-12-r171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tan JC, Patel JJ, Tan A, et al. Optimizing comparative genomic hybridization probes for genotyping and SNP detection in Plasmodium falciparum. Genomics. 2009;93:543–50. doi: 10.1016/j.ygeno.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kidgell C, Volkman SK, Daily J, et al. A systematic map of genetic variation in Plasmodium falciparum. PLoS Pathog. 2006;2:e57. doi: 10.1371/journal.ppat.0020057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jennings CV, Ahouidi AD, Zilversmit M, et al. Molecular analysis of erythrocyte invasion in Plasmodium falciparum isolates from Senegal. Infect.Immun. 2007;75:3531–8. doi: 10.1128/IAI.00122-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jensen JB, Trager W. Plasmodium falciparum in culture: establishment of additional strains. Am.J.Trop.Med.Hyg. 1978;27:743–6. doi: 10.4269/ajtmh.1978.27.743. [DOI] [PubMed] [Google Scholar]
- 18.Trager W, Tershakovec M, Lyandvert L, Stanley H, Lanners N, Gubert E. Clones of the malaria parasite Plasmodium falciparum obtained by microscopic selection: their characterization with regard to knobs, chloroquine sensitivity, and formation of gametocytes. Proc.Natl.Acad.Sci U.S.A. 1981;78:6527–30. doi: 10.1073/pnas.78.10.6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oduola AM, Milhous WK, Weatherly NF, Bowdre JH, Desjardins RE. Plasmodium falciparum: induction of resistance to mefloquine in cloned strains by continuous drug exposure in vitro. Exp.Parasitol. 1988;67:354–60. doi: 10.1016/0014-4894(88)90082-3. [DOI] [PubMed] [Google Scholar]
- 20.Mu J, Awadalla P, Duan J, et al. Recombination hotspots and population structure in Plasmodium falciparum. PLoS Biol. 2005;3:e335. doi: 10.1371/journal.pbio.0030335. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.