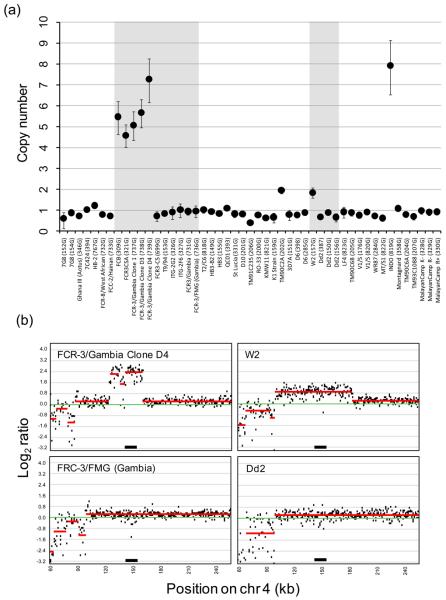
Figure 1. Copy number amplification at PfRh1.
(a) Real-time CN estimates for laboratory strains. The error bars show 95% confidence intervals around estimates of CN. The grey boxes shows strains in the FCR3 or W2/Dd2 group that are genetically identical for microsatellites (this study) and SNPs [12], but differ in PfRh1 CN. Details of the parasites lines used and microsatellite genotypes are shown in Supplementary Table S1. (b) Microarray validation of real-time PCR. Log2 plots of chr 4 for closely related parasites with and without amplification at PfRh1. Points show average Log2 ratios of probes over 500bp windows, while red lines show CN estimates of genome blocks. The two left hand panels show plots for FCR-3/Gambia Clone D4 (7.2 copies by real-time PCR) and FRC-3/FMG (Gambia) (1 copy); the right hand panels show W2 (two copies) and Dd2 (one copy). The amplified region of FCR-3/Gambia Clone D4 and W2 contain 4 and 14 genes respectively and are consistent with a previous report [15]. The position of PfRh1 is shown by a black bar.