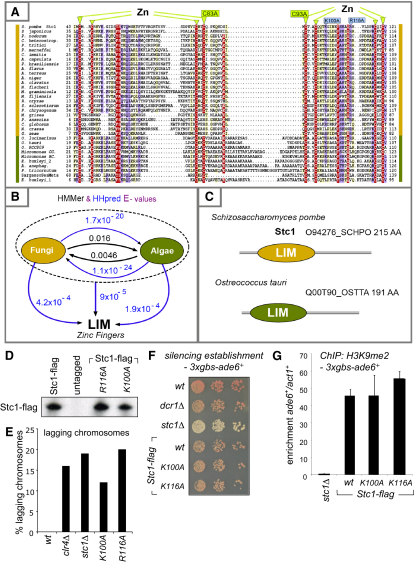
Figure S5.
Stc1 Contains a LIM Domain Required for Its Function, Related to Figure 6
(A) Representative multiple sequence alignment of the N-terminal conserved region of Stc1 homologous proteins. Mutated residues mentioned in the text (C83A, C93A, K100A and R116A) are highlighted. The phyletic groups of Stc1 homologous proteins are indicated by colored bars at each side of the alignment: yellow (fungi) and green (algae). The alignment coloring scheme provides an indication of average BLOSUM62 scores (correlated with amino acid conservation) for each alignment column: red (greater than 3), violet (between 3 and 1) and light yellow (between 1 and 0.3). Sequences were obtained from UniProt, GenBank and DOE-JGI (Joint Genome Institute) databases, but were supplemented by manually assembled ESTs and FGENESH+ predicted gene models (Softberry). Sequences are named according to their species abbreviation. Database of origin, accession numbers (“mod” prefix identifies UniProt sequences corrected by gene prediction software FGENESH+) and full species names are: In fungi: O94276_SCHPO Stc1, Schizosaccharomyces pombe; B6K2E2_SCHJY, SchizoSaccharomyces japonicus; modQ0UKA2_PHANO, Phaeosphaeria nodorum; jgi_Coche, Cochliobolus heterostrophus; B2WPU1_PYRTR, Pyrenophora tritici-repentis; B6QC22_PENMQ, Penicillium marneffei; Q1DX71_COCIM, Coccidioides immitis; modA6RAF6_AJECN, Ajellomyces capsulata; modC1GI81_PARBR, Paracoccidioides brasiliensis; B8NGM5_ASPFN, Aspergillus flavus; Q0CYE9_ASPTN, Aspergillus terreus; modA2QC11_ASPNC, Aspergillus niger; modA1C6Z3_ASPCL, Aspergillus clavatus; A1DHM0_NEOFI, Neosartorya fischeri; jgi_Mycgr, Mycosphaerella graminicola; jgi_Mycfi, Mycosphaerella fijiensis; est1_Rhior EE004662, Rhizopus oryzae; A7EYF8_SCLS1, Sclerotinia sclerotiorum; B6GWR5_PENCW, Penicillium chrysogenum; modA4RHU1_MAGGR, Magnaporthe grisea; modB2B0Z8_PODAN, Podospora anserina; Q2GNQ5_CHAGB, Chaetomium globosum; Q96U02_NEUCR, Neurospora crassa;UPI_GIBZE UPI000023D198, Gibberella zeae. In algae: jgi_Ostlu, Ostreococcus lucimarinus; Q00T90_OSTTA, Ostreococcus tauri; jgi_OstRC, Ostreococcus RCC809; jgi_MicCC, Micromonas CCMP1545; C1E4D8_9CHLO, Micromonas sp. RCC299; jgi2_Emihu, Emiliania huxleyi; jgi_Auran, Aureococcus anophagefferens; modB7GAR1_PHATR, Phaeodactylum tricornutum; SargassoSeaMeta AACY022272749, Sargasso Sea Metagenome; jgi1_Emihu, Emiliania huxleyi.
(B) The N terminus of Stc1 bears statistically significant similarity to LIM domains. Numbers correspond to global profile-to-sequence (in black) and profile-to-profile (in blue) comparison E-values obtained from HMMer and HHpred, respectively (Eddy, 1996; Söding et al., 2005). Arrows indicate the profile search direction.
(C) Domain architectures of two members of the Stc1 family.
(D) Stc1K100A and Stc1R116A LIM domain mutant proteins are stable. FLAG-tagged wild-type and mutant Stc1 proteins were affinity purified from cell lysates and analyzed by western using anti-FLAG antibody.
(E) Stc1 LIM domain mutants exhibit high rates of lagging chromosomes consistent with defects in centromere silencing.
(F) Assay for silencing of 3xgbs-ade6+, mediated by tethered Clr4 (GBD-Clr4-Δcd). The GBD-Clr4-Δcd and 3xgbs-ade6+ constructs were combined in either wild-type or mutant backgrounds to assess establishment of silencing upon recruitment of GBD-Clr4. Establishment of silencing at 3xgbs-ade6+ results in red colonies on limiting adenine; if silencing is not established colonies remain white.
(G) ChIP analysis of H3K9me2 levels on 3xgbs-ade6+.
Eddy, S.R. (1996). Hidden Markov models. Curr. Opin. Struct. Biol. 6, 361–365.
Söding, J., Biegert, A., and Lupas, A.N. (2005). The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33 (Web Server issue), W244–W248.