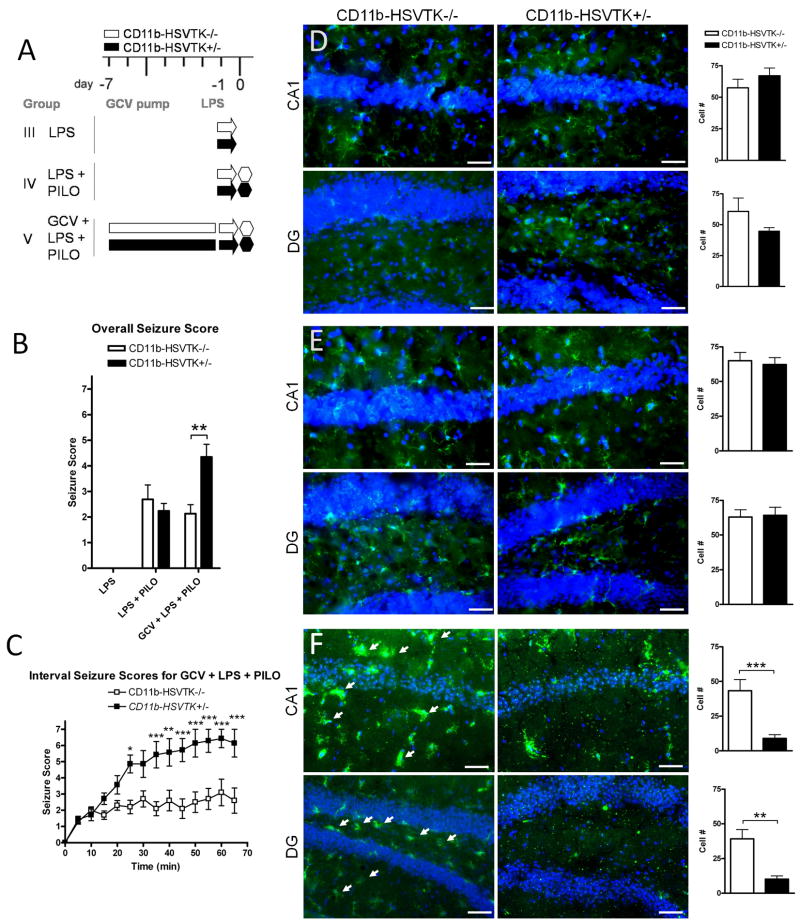
Figure 2. Preconditioning microglia with LPS affects seizure severity differently in CD11b-HSVTK−/− and CD11b-HSVTK+/− mice.
A) Schematic of experimental design in group III: LPS (arrows designate lipopolysaccharide [LPS] only, 1 mg/kg i.p.), IV: LPS + PILO (LPS given 24 hours prior to piloarpine [PILO] injection 280 mg/kg i.p.), and V: GCV + LPS + PILO (gancyclovir [GCV] infusion, LPS given at day −1, then pilocarpine given 24 hours later, 280 mg/kg i.p.). Each genotype is represented, white bars for CD11b-HSVK−/− (wild type mice lacking the transgene) and black bars for CD11b-HSVTK+/− (transgenic mice susceptible to microglia ablation by GCV). Administration of pilocarpine is denoted by a hexagon at day 0 (and is always preceded 15–30 min by methylscopolamine, 2 mg/kg i.p.). Infusion of GCV (for 7 days which continues throughout the experiment) is denoted by the rectangular bars from day −7 to 0. B) Following pilocarpine injection, seizure symptoms were continuously recorded; the maximum score during each 5 minute interval was listed over the 60 minute observation period for each animal and averaged by genotype and group providing the overall seizure score. C) The maximum score during each 5 min interval was plotted over time, data points represent the average score ± S.E.M. for each set of animals. D–F) Tissue was collected 2 hours post seizure induction and stained with Iba1 (green fluorescence) for microglia/macrophages. Iba1 is upregulated in activated cells. CA1 and hilar region of the dentate gyrus (DG) are shown with nuclei (DAPI). Panels in D represent group III, E group IV, and F group V. Arrows in F indicate activated microglia/macrophages. The wild type DG panel in F was taken at slightly different coordinates (a more medial aspect of the granule cell layer). To the right of each set of panels is quantification of Iba1+ cell number counted at 40× in regions of interest (ROIs) sized 300×400μm2 for each group (scale bar 50 μm, two-way ANOVA, Bonferroni post tests, * = p<0.05, ** = p<0.01, *** = p<0.001, data in all graphs plotted as average ± S.E.M.).