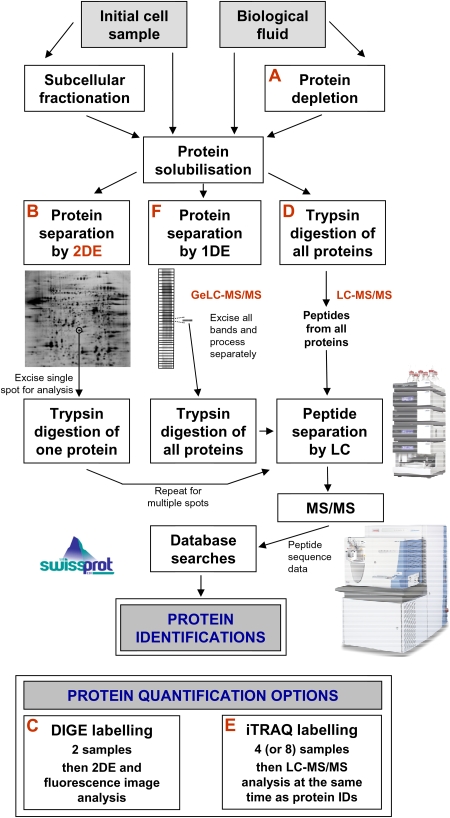
Fig. 1.
Strategies for the global identification and quantification of proteins. Proteins may be derived from biological fluids or solubilized from entire cells or their subcellular fractions. One option with biological fluids is to use proprietary immunodepletion methods (A) to remove up to 20 of the most abundant proteins in order to more readily analyse the lower abundance proteins. Following solubilization, proteins may be separated by 2DE (B). In this workflow, an individual separated protein is removed as a gel plug, trypsin digested and the resulting peptides are typically separated on the basis of relative hydrophobicity by nanoscale LC. The amino acid sequence of these peptides is then determined by MS/MS [19,20] and this data is used to search existing protein databases to achieve a match and, therefore, a protein identification (ID). One can process multiple gel plugs to identify many of the 2DE-separated proteins. The method of choice for 2DE protein quantification is DIGE (C) whereby samples are labelled with different fluorescence dyes prior to mixing together (multiplexing) and 2DE [16]. Such gel-based proteomics remains popular, but for global analysis, it is now more commonplace to trypsin digest the entire solubilized protein mixture to produce a peptide ‘soup’ of all the proteins in the sample (gel-free LC-MS/MS proteomics) (D). Peptides are then separated by LC on the basis of relative hydrophobicity and often also charge as a multidimensional separation. Then, extensive MS/MS and database searches are performed to identify many (ideally all) of the proteins in the original sample. One of the advantages of this workflow is that it is possible to achieve simultaneous quantitative data by introducing an iTRAQ labelling step (E) (or other labelling step) following trypsin digestion without the need for additional LC-MS/MS [17,18]. A further method for global protein identification is to first separate proteins by one-dimensional electrophoresis (1DE) before subjecting individual protein bands to digestion and LC-MS/MS (the geLC-MS workflow) (F). Whilst this is an excellent workflow for high numbers of protein IDs, it is not particularly amenable to protein quantification methodologies. Finally, there are many other options. For example, one may enrich for peptides or proteins of a particular type to study a particular group of proteins and this is most commonly performed for phosphorylation analysis. Peptide isoelectric focussing (IEF) as an additional step within the usual LC-MS/MS workflow is a valid option for increased numbers of IDs and protein LC can also be performed prior to trypsin digestion. Finally, SELDI-TOF MS ProteinChip technology may be potentially used for biomarker discovery, although the approach has certain limitations [21].