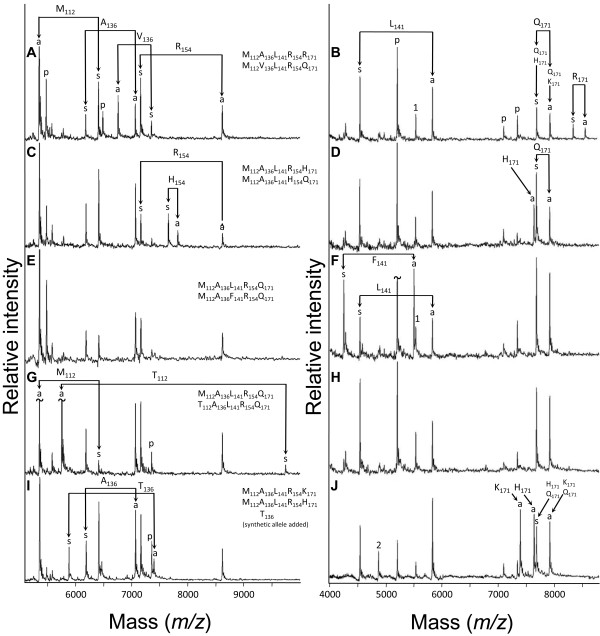
Figure 4.
Mass spectrograms of ovine PRNP codons at positions 112, 136, 141, 154, and 171. A single PCR reaction was used to amplify a 336 bp genomic DNA region and the product split for use in two subsequent multiplex hME reactions. Spectral peaks represent singly-charged ions whose mass-to-charge ratio (m/z) was compared with calibrants for mass determination. Spectra feature labels: s and a, sense and antisense analytes produced from respective hME extension primers; p, unincorporated extension primer; ~, peak height clipped to conserve space. Two artifact peaks are produced as a consequence of multiplex design considerations. The first is a g nucleotide "pausing peak" in the codon 141 antisense assay (5530 Da, feature label "1"). The second artifact peak (feature label "2") is a g nucleotide misincorporation/insertion followed by a ddT termination in the codon 141 sense assay, i.e. 5'-[primer]-CGddT-3' (4866 Da). The correct termination product is 5'-[primer]-CddT-3' (4537 Da). This artifact peak at 4866 Da appears sporadically and independent of sample type or quality. Panels A and B: mass spectrograms illustrating the A136V and Q171R heterozygote. Panels C and D: mass spectrograms illustrating the R154H and Q171H heterozygote. Panels E and F: mass spectrograms illustrating the L141F heterozygote. Panels G and H: mass spectrograms illustrating the M112T heterozygote. Panels I and J: mass spectrograms illustrating the A136T and H171K heterozygote. The T136 was a synthetic allele that was added to the primer extension reaction cocktail to reference animal 200665213 (homozygous for A136).