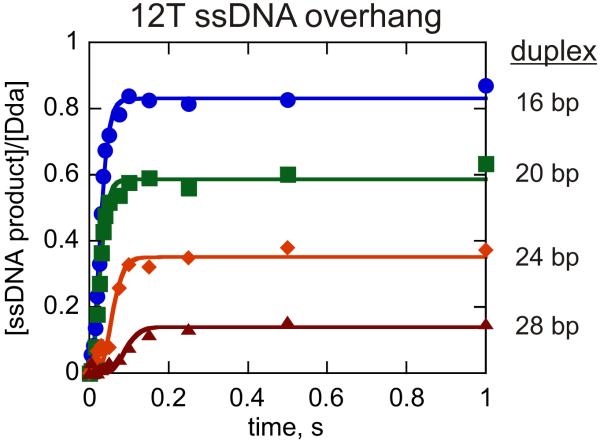
Figure 1.
DNA unwinding by monomeric Dda. Unwinding was measured under conditions in which substrate concentration (100 nM) exceeded the helicase concentration (25 nM) to ensure that only one molecule of Dda was bound per substrate molecule. DNA substrates contained a 12nt overhang of ssDNA and varying lengths of duplex including 12T16bp ( ), 12T20bp (
), 12T20bp ( ), 12T24bp (
), 12T24bp ( ), and 12T28bp (
), and 12T28bp ( ). The experiments were performed in the presence of 5 μM polydT to create single-turnover conditions. The data were fit to equation 5 using Scientist and the resulting kinetic parameters are listed in Table 2.
). The experiments were performed in the presence of 5 μM polydT to create single-turnover conditions. The data were fit to equation 5 using Scientist and the resulting kinetic parameters are listed in Table 2.