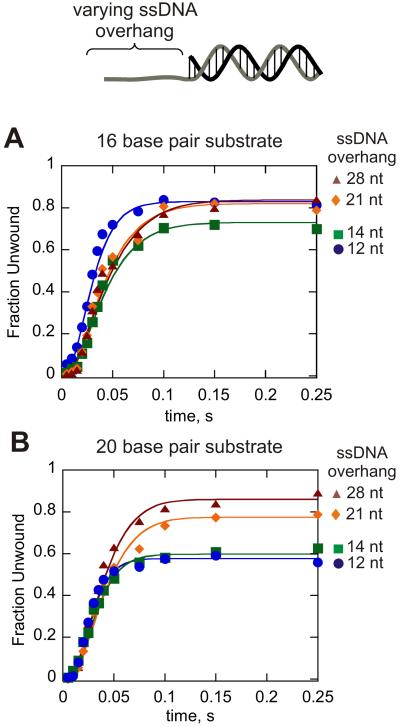
Figure 3.
Dda helicase-catalyzed unwinding of 16 bp (A) and 20 bp (B) substrates containing varying length ssDNA overhang. For all substrates containing 12 nt of ssDNA, the DNA substrate concentration (100 nM) exceeded the concentration of Dda (25 nM) and the quantity of product was divided by the enzyme concentration to obtain the fraction unwound. These conditions were chosen to ensure that only one molecule of Dda was bound to the substrate. For all other substrates, the concentration of Dda (100 nM) exceeded the DNA substrate concentration (10 nM) in order to ensure that multiple molecules were bound to the ssDNA overhangs. All experiments were performed in the presence of 5 μM poly dT to create single-turnover conditions with respect to the DNA substrate. The lengths of the ssDNA overhangs are listed in the figure. Data were fit to a model as depicted in Figure 2 by using the program Kintek Global Kinetic Explorer (Kintek Corp.). Three or four kinetic steps for unwinding were used for the unwinding step with the 16 bp substrate and 20 bp substrate, respectively. One, two, three, or four molecules of Dda were pre-bound to the DNA substrate for the 12 nt, 14 nt, 21 nt, and 28 nt substrates, respectively. The rate constant for translocation was set equal to that for DNA unwinding. The rate constants for DNA unwinding, ku, and dissociation, kd, were allowed to float for each substrate to obtain the best fit for each mechanism. The resulting rate constants are shown in Table 3.