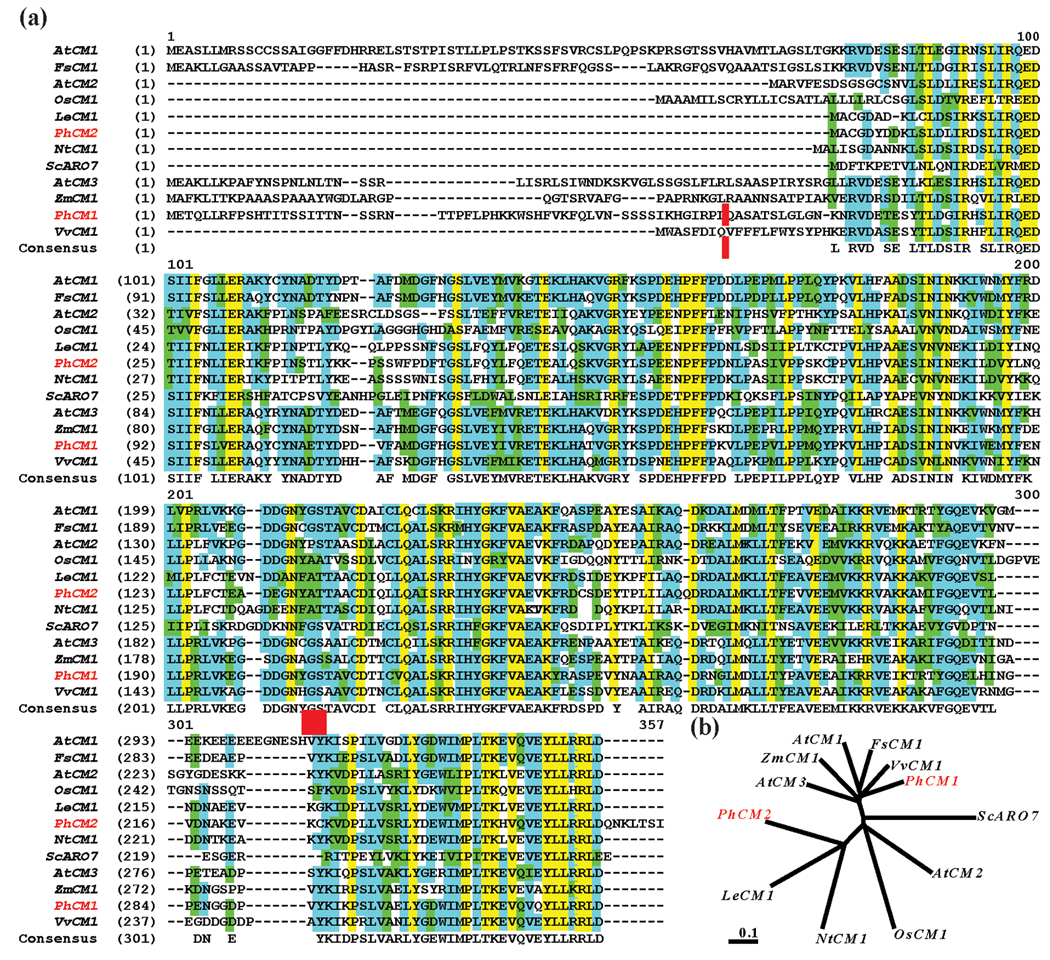
Figure 2.
Predicted peptide sequence alignment and an unrooted neighbor-joining phylogenetic tree of CM proteins from various species. Sequences represented are from Arabidopsis thaliana (accession: NP_566846, NP_196648, and NP_177096), Fagus sylvatica (ABA54871), Solanum lycopersicum (AAD48923), Nicotiana tabacum (BAD26595), Oryza sativa (NP_001061910), Petunia x hybrida (EU751616, EU751617), Vitis vinifera (CAO15322), Zea mays (AY103806), and Saccharomyces cerevisiae (NP_015385). (a) Sequences were aligned using the AlignX program of the Vector NTI Advance 10.3.0 software (Invitrogen). Residues highlighted in: blue represent consensus residues derived from a block of similar residues at a given position, green represent consensus residues derived from the occurrence of greater than 50 % of a single residue at a given position, and yellow represent consensus residues derived from a completely conserved residue at a given position. Petunia sequences are highlighted in red to the left, a red vertical bar represents the beginning of the mature protein sequence used for PhCM1, and a red box indicates an allosteric regulatory site (GS). (b) TREEVIEW software with the nearest-joining method was used to create the resulting tree. Scale bar represents distance as the number of substitutions per site (i.e., 0.1 amino acid substitutions per site).