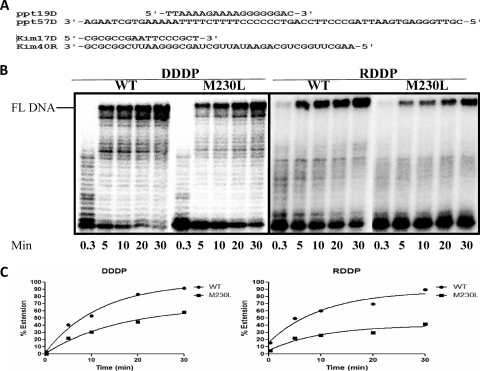
FIG. 4.
Processive polymerization by WT and M230L mutant RTs. (A) Graphic representation of the T-P systems used to monitor the DDDP and RDDP activities of recombinant RTs. DNA primers ppt19D and kim17D were labeled with 32P and annealed to DNA template ppt57D and RNA template kim40R, respectively. (B) Polymerization activities were analyzed by monitoring the percentage of full-length (FL) DNA synthesis in time course experiments in the presence of the heparin trap. The position of full-length DNA is indicated on the left side of the panel. All reactions were resolved by denaturing 6% polyacrylamide gel electrophoresis, visualized by phosphorimaging, and quantified with ImageQuant software (GE Healthcare). (C) Graphic representation of the calculated percentage of full-length DNA synthesis in the time course experiments from the gel results. Band intensities were calculated by ImageQuant software, and the data were curve fit by GraphPad Prism (version 5.01) software.