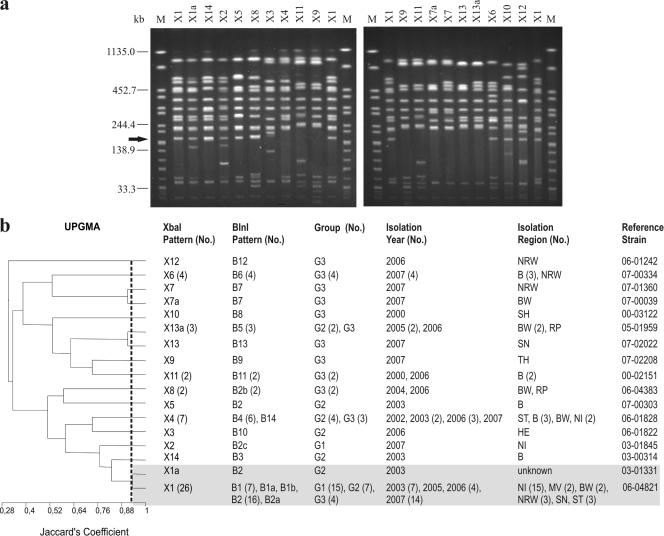
FIG. 1.
(a) XbaI PFGE profiles of representative S. Saintpaul isolates. To define the profiles, only bands at >33 kb were considered. Similar profiles with only a one-band difference were designated by letters. Profiles with differences in two or more bands were designated by numbers. Profiles X1 and X1a are the profiles of the clonal line. The arrow indicates the position of the aadB gene. Lanes M contained XbaI-digested DNA of S. enterica serovar Braenderup H9812, which was used as a size standard. (b) Dendrogram showing the genetic similarity between XbaI profiles determined by using the unweighted-pair group method with arithmetic averages (UPGMA) and Jaccard's coefficient (J). The numbers in parentheses are the numbers of isolates when there was more than one isolate. G1, group 1 (baseline study); G2, group 2 (diagnostic feces study); G3, group 3 (diagnostic food study). Abbreviations for isolation regions in Germany are explained in Table 1. The shading indicates related clonal patterns at a Jaccard's coefficient of 0.93.