FIG. 5.
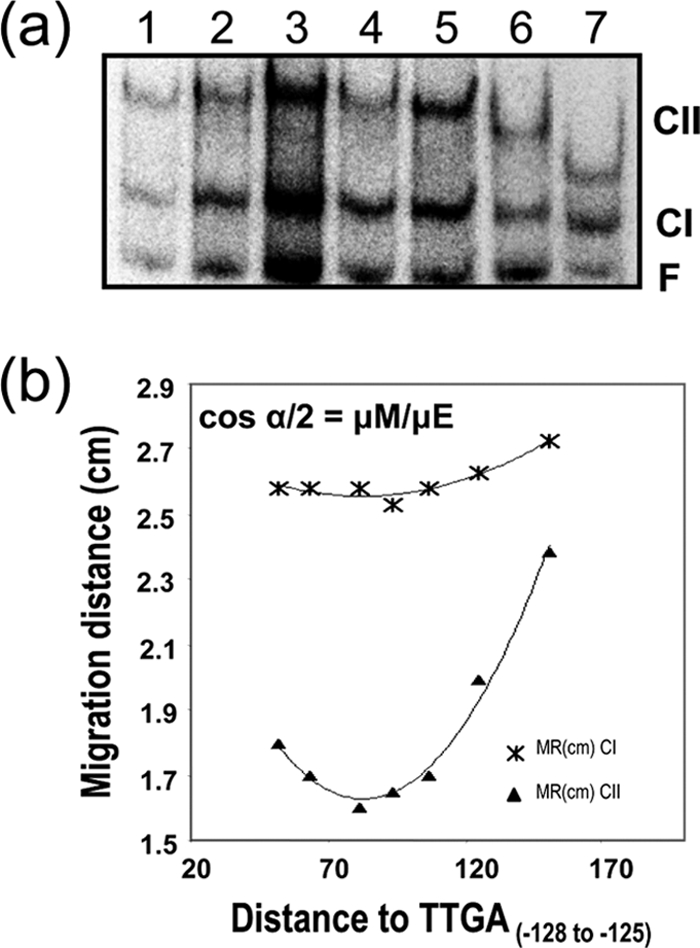
XylS-CTD binding to DNA provokes DNA bending. Gel retardation assays of XylS-CTD on a series of 195 bp DNA fragments, containing XylS binding sites in permutated locations were run in 8% polyacrylamide (wt/vol). The migration distance of each complex (*, CI; ▴, CII) was plotted against the distance between XylS binding site 5′ end and fragment 5′ end in each fragment tested. The bending angle for each complex was estimated according to the equation cos α/2 = μM/μE, where μM is the migration of the complex with lowest mobility and μE is the migration of the complex with highest mobility. Values are the average of six independent determinations.
