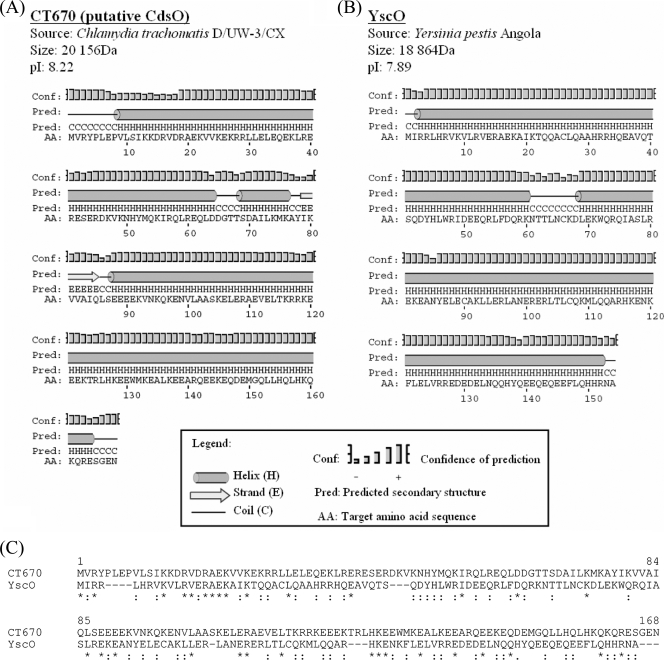
FIG. 4.
Shared protein characteristics of CT670 and YscO. (A and B) Protein, source, size, pI, and predicted secondary structure for C. trachomatis D/UW-3/CX (A) and Y. pestis Angola (B). Secondary structure predictions were obtained with the PSIPRED algorithm using the primary protein sequences of CT670 and YscO (30). (C) Sequence alignment of CT670 and the YscO protein from Y. pestis. The alignment was constructed using the ClustalX 1.83 program. Identical residues are indicated by asterisks, and conservative substitutions are indicated by colons. The significance of sequence similarity between the two proteins was determined using the PCOMPARE program of the PCGENE software package (39) with the unitary matrix and a gap penalty value of 3. The YscO sequence was randomized 100 times. The normal score of this sequence alignment differed from the mean score of the randomized sequence alignment by 3.7 standard deviations, indicating that the observed sequence similarity of CT670 and YscO is significant and it is not due to the amino acid compositions of the proteins by chance.