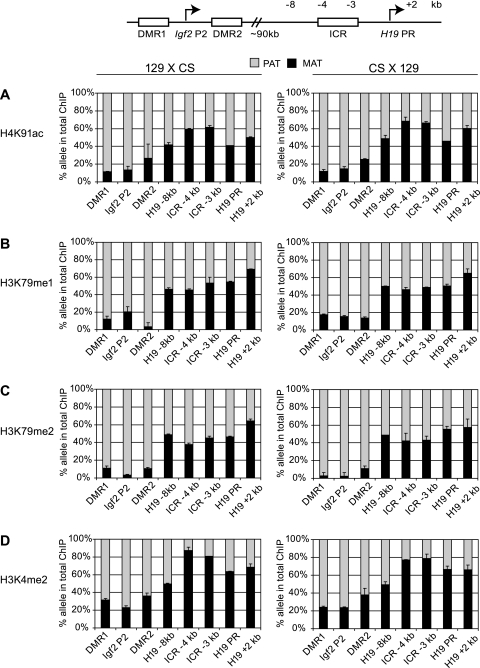
FIG. 1.
Activating chromatin composition along the H19/Igf2 imprinted domain. Allele-specific activating chromatin was measured by quantitative ChIP-SNuPE assays at the H19/Igf2 imprinted domain, using the 7-plex assay (see Fig. S1 in the supplemental material) and the H19 promoter assay. The regions of interest are depicted in the schematic drawing and indicated under each column. ChIP was done in duplicate, using antibodies against specific histone modifications (indicated on the left side of each row of charts) to precipitate chromatin from 129 mother × CS father MEFs or reciprocal CS mother × 129 father MEFs (indicated at the top). The ratio of an allele-specific histone modification at a specific region was expressed as a percentage of maternal (MAT) or paternal (PAT) allele in the total (maternal plus paternal, or 100%) immunoprecipitation. Standard deviations are indicated as error bars. Active chromatin histone globular domain modifications H4K91ac (A), H3K79me1 (B), and H3K79me2 (C) and the control histone tail modification H3K4me2 (D) clearly distinguished the paternal alleles at the Igf2 regions. These modifications were slightly biased or not biased toward the maternal alleles at the H19 regions. No allele-specific chromatin differences existed at a “neutral” intermediary region −8 kb upstream of the H19 promoter (PR). Reciprocal mouse crosses had very similar allele-specific chromatin composition.