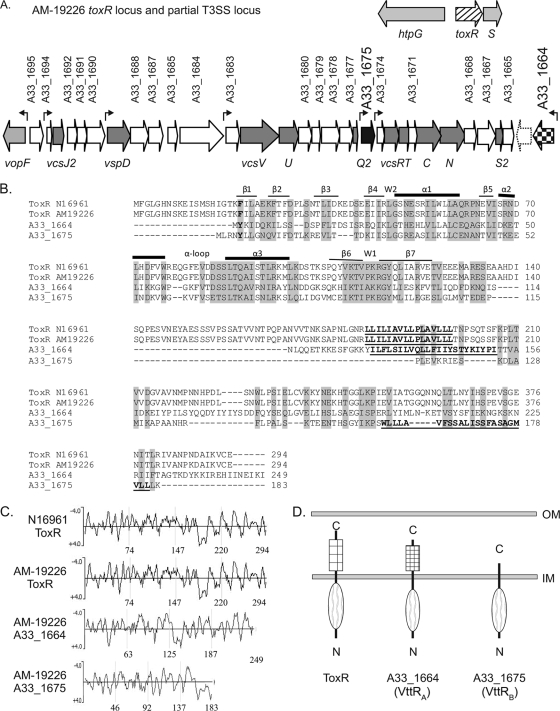
FIG. 1.
(A) The flanking genes for the ancestral toxR gene (hatched arrow) and the region of the T3SS pathogenicity island encoding the structural components (dark-gray arrows) and the two putative transcriptional regulators (black and checkered arrows) are shown. White arrows represent genes encoding hypothetical or conserved hypothetical proteins. The dotted arrow represents a gene present in our annotation but not annotated by the J. Craig Venter Institute. Genes encoding known proteins are shown in light gray. The seven small arrows above the genes indicate the locations of predicted promoter sequences. (B) Multiple-sequence alignment (ClustalW2 with default settings) of the ToxRN16961, ToxRAM-19226, A33_1664, and A33_1675 protein sequences. Amino acid residues that constitute the predicted transmembrane domains are in bold and underlined. The predicted secondary structures of the ToxR domains comprising the winged HTH motif are indicated above the N16961 ToxR sequence. Residues forming beta sheets are indicated by thin lines, those forming alpha helices are indicated by thick lines, and wing residues are indicated by the letter W. (C) Hydrophilicity plots of ToxR and the ToxR-related proteins using Kyte-Doolittle analysis. Hydrophilic residues have a negative score and hydrophobic residues have a positive score on the plot. The numbers at the bottom of each panel refer to amino acid positions within the four proteins. (D) Domain structure and membrane localization of ToxR paralogs based on TMHMM analysis, hydrophilicity plots, and phoA fusion analysis. OM, outer membrane; IM, inner membrane.