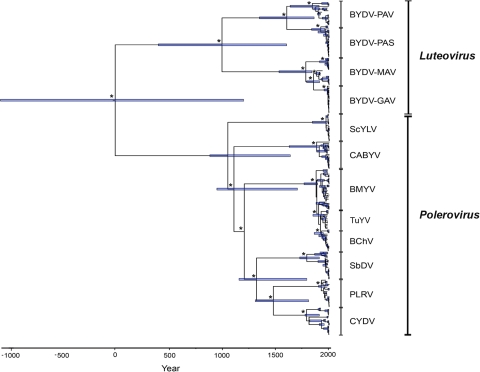
FIG. 2.
Maximum clade credibility phylogeny of the family Luteoviridae based on the CPov data set using 20 sequences per species and an empirical prior distribution on the substitution rate. Branch tip times reflect the times of viral sampling. The tree is automatically rooted through the use of a relaxed molecular clock, and the total depth of the tree is the TMRCA of the family Luteoviridae. Asterisks indicate nodes with posterior probabilities of ≥0.90. Horizontal blue bars denote 95% HPD intervals for the age of each node. Vertical black bars delimit sequence clusters for each virus genus and species.