FIG. 4.
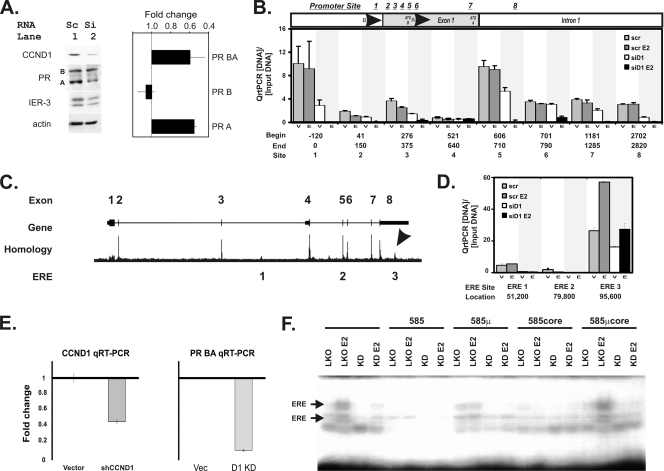
Small interfering RNA (siRNA) knockdown of cyclin D1 blocks progesterone receptor expression and inhibits chromatin binding of the estrogen receptor to a novel 3′ estrogen response element in the progesterone receptor gene. (A) A small interfering RNA that knocks down cyclin D1 (siCCND1) decreases progesterone receptor expression. T47D breast cells were transfected with siCCND1 (Si) or a scramble control (Sc), and protein lysates were harvested 48 h later (left panel). We performed immunoblot analyses using 50 μg of protein lysates for the indicated genes. Immunoblots for cyclin D1 (CCND1), the progesterone receptor (PR) isoforms A and B, the immediate-early response gene 3 (IER-3) that we had previously demonstrated to have responded to cyclin D1 and estrogen (48), and an actin loading control are shown. (Right panel) Isoform BA and B transcripts of the progesterone receptor were measured using equal amounts of reverse-transcribed cDNA via isoform-specific qRT-PCR. Isoform A levels were derived by subtracting B from AB levels. The mean and standard error fold changes from scramble to siCCND1 transfectants are shown. (B) Analysis of cis sequences that have been reported to be estrogen control elements in the progesterone receptor gene promoters. Sites in the progesterone promoter region where estrogen regulation of PR expression has been proposed to occur are indicated. Candidate sites include the following: site 1, Sp1 (39); site 2, an AP1 at +90 from the PR B transcription start site (TSS) (33); site 3, a G/A 326 GATA5 breast cancer risk polymorphism (17); site 4, an Sp1-ERE half-site (33); site 5, the PR isoform A promoter region; site 6, a second AP1 at +745 from the PR B TSS (38); site 7, an RNA polymerase II binding site (6); and site 8, a rat promoter site that was previously shown to have responded to E2 (20). The locations of the start (Begin) and ending (End) points for the PCR primers used in these studies are indicated, along with the numbering system (Site) that identifies each site. We performed ChIP-PCR for ER binding as previously described (48). Anti-ER ChIP-PCR signals are plotted as a fraction of the input DNA for each reaction, subtracting the minimal ChIP-PCR backgrounds obtained using a nonspecific control serum from the αER signal. We measured αER ChIP-PCR DNA levels in cells transfected with a scramble control (scr) or an siCCND1 (siD1) oligonucleotide. Transfectants were treated with vehicle control (V) or estradiol (E). At each site, the bars depict the αER signal for scramble plus vehicle (scr), scramble plus E2 (scr E2), siCCND1 plus vehicle (siD1), and siCCND1 plus E2 (siD1 E2). (C) Locations of three conserved, canonical estrogen receptor binding sequences in the progesterone receptor gene. The progesterone receptor gene structure is shown diagrammatically using the UCSC genome browser to plot its exons. A homology plot from the genome browser is shown; an arrow identifies the region in the 3′ UTR of the gene that is the most highly conserved part other than its exons. The locations of the three canonical estrogen receptor binding elements that are conserved across mammalian species are indicated (ERE 1, 2, and 3). (D) Chromatin immunoprecipitation (ChIP) identified a novel ER-binding site in the DNA encoding the 3′ untranslated region (UTR) of the PR gene whose ER binding is uniquely stimulated by estradiol and blocked by an inhibitory siCCND1 oligonucleotide. We performed ChIP-PCR for ER binding at the three ERE sites as described for panel C. Anti-ER ChIP-PCR signals are plotted as a fraction of the input DNA for each reaction, subtracting the ChIP-PCR backgrounds. We measured αER ChIP-PCR DNA in cells transfected with a scramble control (scr) or an siCCND1 (siD1) oligonucleotide. Transfectants were treated with vehicle control (V) or estradiol (E). For each site, the bars depict the αER signal for scramble plus vehicle (scr), scramble plus E2 (scr E2), siCCND1 plus vehicle (siD1), and siCCND1 plus E2 (siD1 E2). Three conserved, canonical estrogen binding elements identified in the UCSC genome browser were compared; these sites are indicated according to the nucleotide distance downstream of the transcription start site of the B isoform of the progesterone receptor. (E) Stable knockdown of cyclin D1 reduces PR mRNA levels. We stably reduced cyclin D1 expression in T47D cells by expressing a lentiviral shRNA construct in pooled transfectants selected using puromycin. Changes in cyclin D1 and progesterone receptor mRNAs are shown as fold changes in mRNA levels compared to those of pooled control cells containing the parent pLKO vector (Vec). We used qRT-PCR to monitor mRNA levels as described in Materials and Methods. (F) Stable knockdown of cyclin D1 reduces binding to the canonical estrogen receptor binding site in the 3′ ERE of the progesterone receptor gene. We used electrophoretic mobility shift assays to monitor protein binding to an oligonucleotide containing the estrogen response element sequences 95,585 nucleotides downstream of the PR transcription start site. Protein binding to the oligonucleotide probe was evaluated in lysates harvested from the vector control cells (LKO) in the absence and presence of estradiol (E2). Levels of binding to the radiolabeled probe were compared using lysates harvested from the cells expressing the cyclin D1 knockdown shRNA (KD) in the absence and presence of estradiol (E2). The specificity of the binding was evaluated using a 200-fold excess of unlabeled probe (585), unlabeled 585 oligonucleotide containing a mutation in the ERE (585μ), an unlabeled smaller oligonucleotide containing the core ERE sequence (585core), and an unlabeled core oligonucleotide containing the ERE-inactivating mutation (EREμcore).