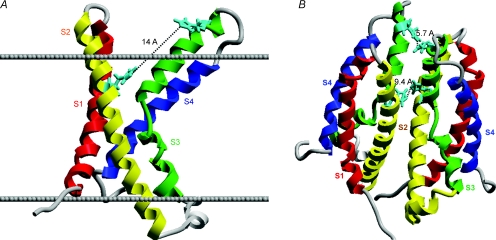
Figure 1. Homology models of the HV1 proton channel as a monomer (A) or a dimer (B), showing the locations of the two His residues exposed to the extracellular solution.
In A, the extracellular surface of the membrane is indicated by the upper line, the intracellular surface by the lower line. His140 and His193 are shown in aqua. In the monomer, His140 and His193 are 14 Å apart, too far to plausibly coordinate a Zn2+ atom. In the dimer (B), the His140 from each monomer are closer together, as are the His193; either or both could comprise high-affinity Zn2+ binding sites. Thus, a high-affinity Zn2+ binding site exists only in the dimer. The recent identification of proton channel genes allows homology-based structural predictions, because the proton channel molecule bears striking homology to the voltage-sensing domain of K+ and other voltage-gated ion channels (Ramsey et al. 2006; Sasaki et al. 2006). HV1 contains four transmembrane domains resembling S1–S4 of other channels, but lacks the S5–S6 regions that comprise the ion conduction pathway in other channels.