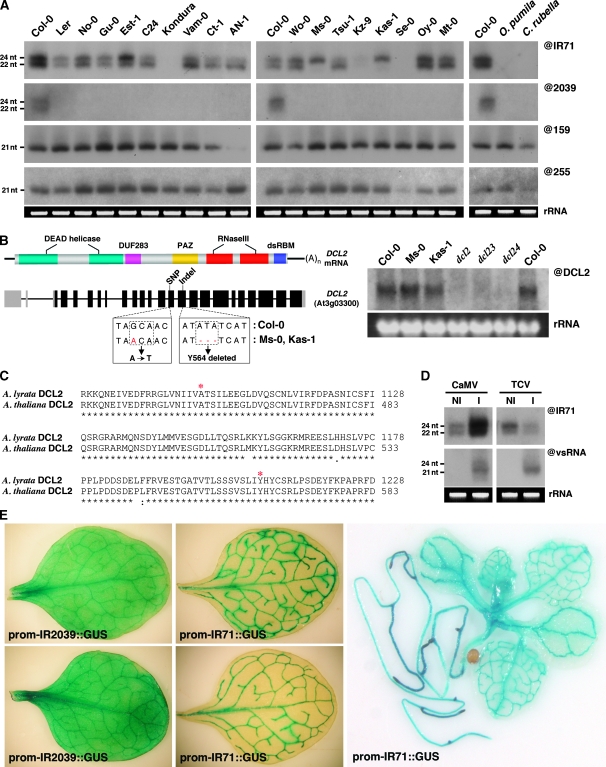
Figure 4.
Rapid evolution and regulated expression of endogenous IR loci. (A) Northern analysis of IR71- and IR2039-derived siRNAs in various Arabidopsis accessions representing the genetic diversity within the A. thaliana species. Also shown are miRNA (@159) and trans-acting siRNA (@255) in each accession. (B) A schematic representation of the DCL2 mRNA showing the domain structure and genomic region with the indicated SNP and indel in ecotypes Ms-0 and Kas-1 (boxed). The right panel shows that the mutations in the DCL2 genomic sequence do not affect mRNA accumulation (@DCL2) in the indicated accessions as assessed by RNA gel blot analysis. (C) Alignment of DCL2 amino acid sequence covering both the predicted amino acid substitution and deletion (marked *) between a representative sequence present in most Arabidopsis accessions (except Ms-0 and Kas-1) and the distinct species A. lyrata. (D) Northern analysis of virus-infected Col-0 plants probed with either IR71- (@IR71) or virus siRNA (vsRNA)-specific probes. NI, non-infected and I, infected with either CaMV (Cauliflower mosaic virus) or TCV (Turnip crinkle virus). (E) GUS staining of leaves from promoter::GUS fusions of two independent transgenic lines containing ∼1.5-kb upstream promoter regions from either IR2039 (left) or IR71 (middle). The right panel shows GUS staining of a whole plant representative of the IR71 promoter::GUS fusion lines.