Figure 1. Expression profiles of TAP-1, TAP-2, and TAP-3.
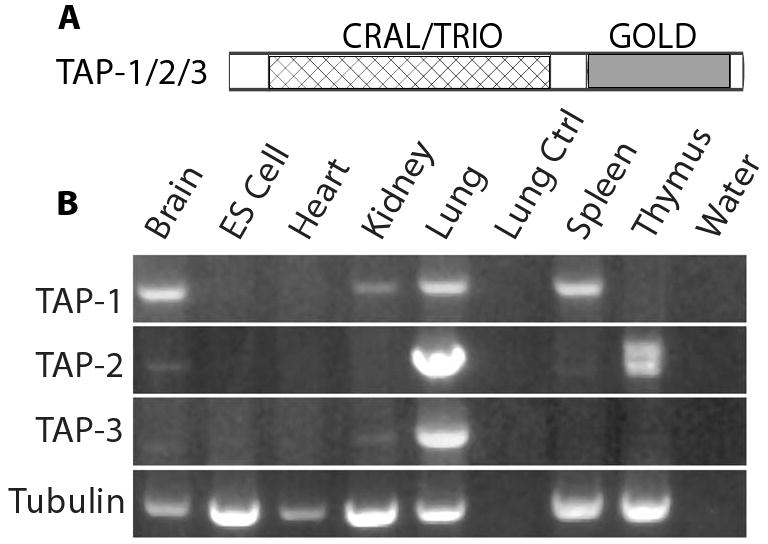
A. The domain structure of TAP proteins. B. RNA purified from mouse tissues (brain, heart, kidney, lung, spleen and thymus) and mouse ES cells was used to generate cDNA. Bands are ethidium bromide stained fragments of DNA that were amplified from the cDNA using primers designed to amplify TAP-1, TAP-2 and TAP-3. Primers that specifically amplified tubulin were used as a control for the input cDNA, and this experiment demonstrated the presence of amplifiable RNA in each tissue sample. To control for non-specific amplification, we analyzed control samples that contained RNA but no cDNA (Lung Ctrl), or contained no RNA (water). TAP-1 was detected in brain, kidney, lung and spleen, TAP-2 was detected in lung and thymus, and TAP-3 was detected only in lung. The TAP-1 and TAP-3 primers generated a single band in all cases, whereas the TAP-2 primers generated two bands from thymus tissue that may result from alternatively spliced transcripts.
