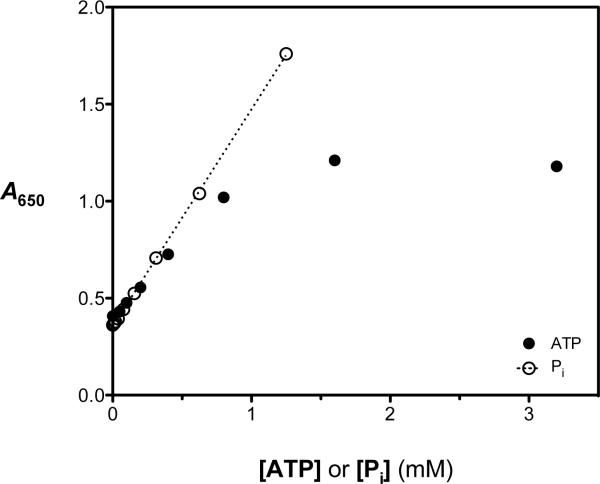
Figure 1.
Evaluation of the dependence of the phosphomolybdate blue (PMB) ATPase assay signal on free phosphate (open circles) and initial ATP (filled circles) concentrations under otherwise optimized conditions. In the presence of RecA (0.5 μM) and poly(dT) (5 μM-nts), the PMB assay reagent produced a linear colorimetric response (absorbance at 650 nm, A650) when free phosphate (Pi; open circles) was added up to 1.25 mM. In separate experiments, the RecA-DNA ATPase reaction was allowed to proceed with various starting ATP concentrations (0.01 – 3.5 mM). After 35 min, the reactions were stopped by addition of the PMB assay reagent and the A650 signal was measured. The A650 values were plotted as a function of initial [ATP] (filled circles), and the results demonstrate that the assay signal is linear with ATP concentration up to 0.8 mM but is substantially nonlinear between 0.8 and 3.2 mM ATP.