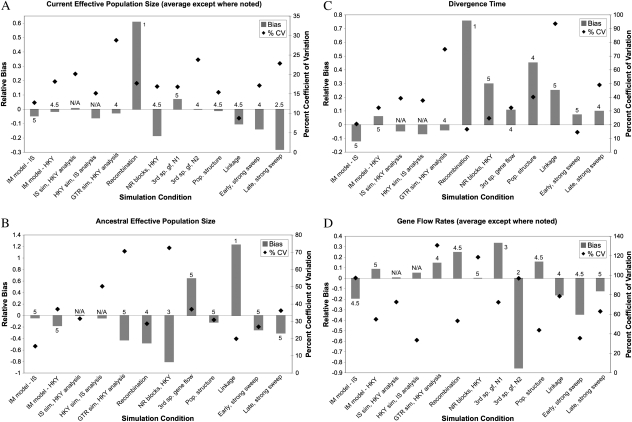
FIG. 1.
Relative bias based on average MLE and coefficient of variation (CV) in MLE values for various simulation conditions in estimates of (A) current effective population size, averaged over the two species (values reported separately for third species gene flow); (B) ancestral effective population size; (C) divergence time; and (D) interspecific gene flow, averaged over the two species (values reported separately for third species gene flow). Bias is calculated as (average MLE − true value)/(true value). For violations with multiple levels of severity, a single intermediate level was used here: ρ = 0.02 per bp for recombination violations, Nefm = 0.3 for third species gene flow, Nefm = 0.5 for population structure, r = 0.02 for interlocus linkage. Data for all levels of severity are shown in supplementary file S3, Supplementary Material online. Numbers above or next to bars are the number of replicates out of five (averaged over the two species or two directions for current Nef and gene flow rates, respectively, except for third species gene flow) for which the HPD interval contains the true value. IS-simulated data analyzed with the HKY model and HKY-simulated data analyzed with the IS model were compared with analyses using the correct model rather than to the true values, so no numbers are given in those cases. Note that bias and CV are plotted on different axes.