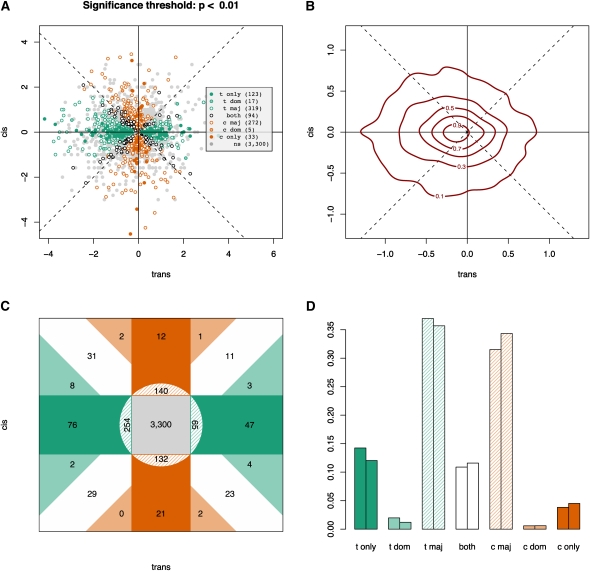
Figure 2.
Genome-wide ASE polymorphism in S. cerevisiae: (A–C) y-axis, log2(ecis), versus x-axis, log2(etrans). (A) Scatter plot of cis and trans estimators with the shading of the points indicating what category individual genes fall into, as determined by their two-dimensional 99% confidence intervals. These classifications are made with respect to the relationship between confidence intervals of each point and the four lines running through the origin, which are the two axes and the two diagonals. Trans-only (t only) and cis-only (c only) indicate that the confidence intervals for the genes, respectively, overlap the log2(ecis) = 0 or the log2(etrans) = 0 axis line only. “Both” refers to the genes that only overlap either the log2(ecis) = log2(etrans) line or the log2(ecis) = −log2(etrans) line (the positive and negative diagonals). Trans-dominant (t dom) and cis-dominant (c dom) genes overlap no lines at all but fall in the quadrants nearest the log2(ecis) = 0 and log2(etrans) = 0 lines, respectively. Trans-major (t maj) overlaps the log2(ecis) = 0 line and at least one of the diagonals, whereas cis-major (c maj) overlaps the log2(etrans) = 0 line and at least one of the diagonals. “ns” indicates nonsignificant genes: each of them overlaps both the log2(ecis) = 0 and the log2(etrans) = 0 lines. A more detailed explanation of how genes are classified can be found in Supplemental Figure S6. (B) A contour plot of the two-dimenstional probability density function of the data (from the two-dimensional kernel density estimator in the MASS library in R) indicating where most genes fall in the cis/trans space using independent estimates of cis and trans. The “elevation” indicated by the contours expresses the probability of a point falling in that region. The total volume beneath the surface sums to unity. (C) A summary of classifications based on the results from Figure 1A. Dark green, trans-only; light green, trans-dominant; dark orange, cis-only; light orange, cis-dominant; hashed regions, the “cis/trans-major” classifications; white, both; gray, nonsignificant genes. (D) The histogram indicates that significant trans changes dominate in comparison to significant cis changes. The left bar of each pair is before discarding the 1265 “hotspot” genes from Yvert et al. (2003), and the right bar is after discarding them. Importantly, trans-dominant and cis-dominant are not to be confused with genetic dominance; instead, they are meant to convey that the magnitude of trans variation is greater than cis variation or the reverse, respectively.