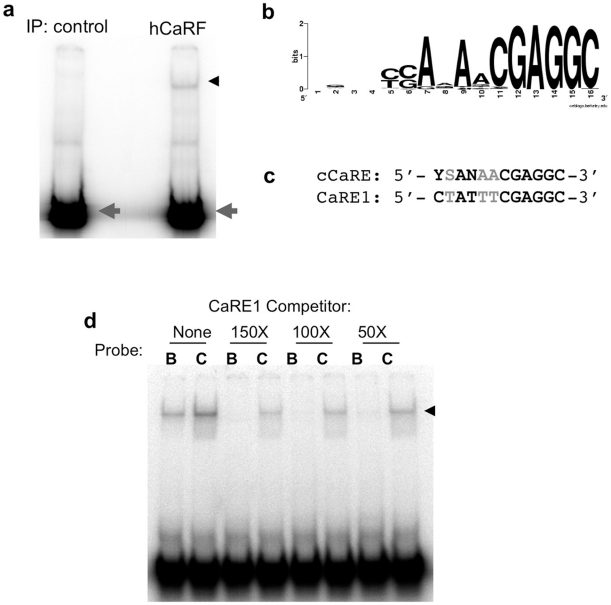
Figure 3. Identification of a consensus CaRF binding element.
hCaRF synthesized by TNT (hCaRF) or control rabbit reticulocyte without CaRF (control) was used to coprecipitate oligonucleotides from a library of random 16mers. a) After four rounds of enrichment and amplification, the final pulldown from each sample was radiolabeled and mixed with hCaRF for evaluation by EMSA. Equal amounts of radiolabeled oligos are present in each pool (gray arrowhead), however a CaRF binding band is retarded only from the pool that was isolated by coprecipitation with hCaRF (black arrowhead). b) WebLogo (http://weblogo.berkeley.edu/) representation of the cCaRE consensus motif derived from the 62 sequences in Table S3. The position of the bases is indicated along the bottom from 1–16, and the height of the letters indicates the enrichment of that base at each position. If all four bases were equally likely to be present at any position, no base is indicated. c) Alignment of the cCaRE and CaRE1 motifs. Black indicates bases that are conserved between the elements, and gray shows bases that vary. Y = C/T, S = C/G, and N = any base. d) Comparison of the affinity of CaRF for CaRE1 and cCaRE. A constant amount of hCaRF was bound to radiolabeled CaRE1 (B) or cCaRE (C) probes and the relative affinity of the interactions were assessed by competition EMSA upon the addition of a 150, 100, or 50-fold molar excess of unlabeled CaRE1 probe. The band retarded upon CaRF binding is indicated by the arrowhead.