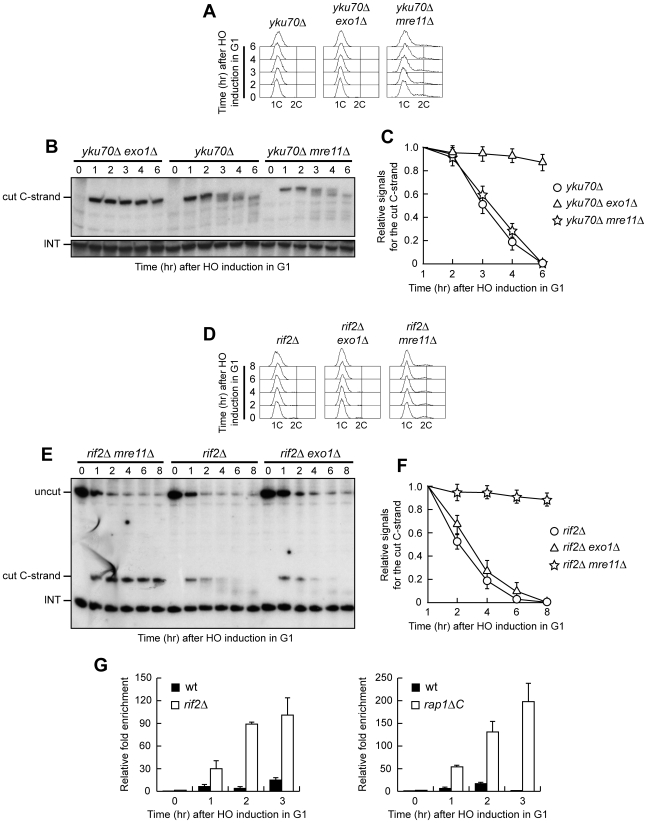
Figure 7. Nuclease requirements for ssDNA generation at a de novo telomere.
(A–C) HO expression was induced at time zero by galactose addition to α-factor-arrested yku70Δ, yku70Δ exo1Δ and yku70Δ mre11Δ cell cultures that were then kept arrested in G1. (A) FACS analysis of DNA content. (B) RsaI-digested genomic DNA was hybridized with probe A as in Figure 3E. (C) Densitometric analysis. Plotted values are the mean value ±SD from three independent experiments as in (B). (D–F) HO expression was induced at time zero by galactose addition to α-factor-arrested rif2Δ, rif2Δ exo1Δ and rif2Δ mre11Δ cell cultures that were then kept arrested in G1. (D) FACS analysis of DNA content. (E) RsaI- and EcoRV-digested genomic DNA was hybridized with probe A as described in Figure 1C. (F) Densitometric analysis. Plotted values are the mean value ±SD from three independent experiments as in (E). (G) HO expression was induced at time zero by galactose addition to α-factor-arrested wild type, rif2Δ and rap1ΔC cells, all expressing a fully functional MRE11-MYC tagged allele. Cells were then kept arrested in G1 and chromatin samples taken at different times after HO induction were immunoprecipitated with anti-Myc antibody. Coimmunoprecipitated DNA was analyzed by quantitative real-time PCR (qPCR) using primer pairs located at the nontelomeric ARO1 fragment of chromosome IV (CON) and 640 bp proximal to the HO site (TEL), respectively. Data are expressed as relative fold enrichment of TEL over CON signal after normalization to input signals for each primer set. The data presented are the mean of those obtained in three independent experiments. Error bars indicate s. d.