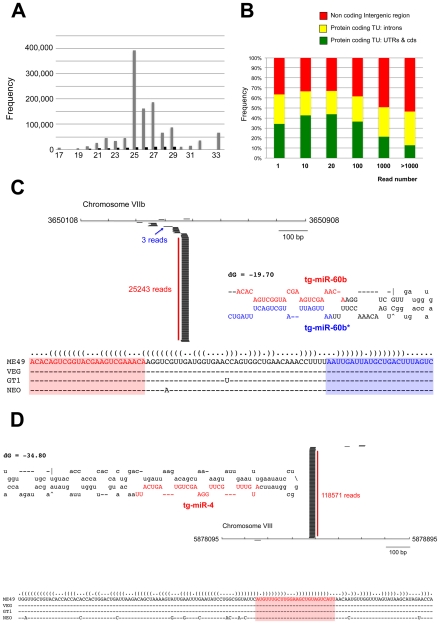
Figure 3. A repertoire of Toxoplasma endogenous small RNAs.
(A) Size distribution of Toxoplasma sRNAs. The sets of redundant (grey) and unique (black) small RNAs were used to generate a histogram quantifying the number of sequences obtained for each size class. nt., nucleotides. (B) Distribution of small RNAs across the Toxoplasma genome. TU: Transcriptional unit, CDS: CoDing Sequence, UTRs: UnTranslated Regions. (C) A miR-60b production hot spot in chromosome VIIb is shown. Small RNAs with perfect matches were plotted within a 800-bp sliding window. Short thin lines above the long bars represent small RNAs derived from the antisense strands, and lines below the bars represent small RNAs from the sense strands. Vertical bars represent the consensus positions of sequencing reads that mapped to the predicted precursors, and the values indicate the total number of these reads. The fold-back structure of the precursor predicted with mfold is provided below. The mature region is shown in red and the passenger strand (microRNA*) in blue. The miR-60b–containing locus for the three canonical strains of Toxoplasma and Neospora caninum is depicted by sequence and structure. (D) A miR-4 production hot spot in chromosome VIII is shown along with the predicted structure and the sequence conservation across parasite species.