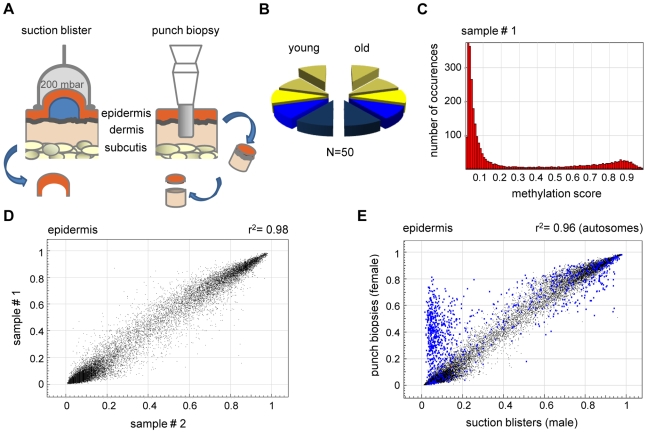
Figure 1. High interindividual similarity between DNA methylation patterns from human epidermis.
(A) Procedures for skin sample preparation. Suction blisters were induced on the forearms of healthy volunteers and suction blister roofs were taken as sources for epidermal DNA (left panel). Punch biopsies were obtained from the outer forearm (sun-exposed) and inner arm (sun-protected) and separated into epidermal and dermal layers by dispase II treatment. (B) Schematic outline of the sample groups analyzed in this study. Epidermis (yellow) and dermis (blue) samples were obtained from sun-exposed (bright colors) and sun-protected (shaded colors) skin areas, either as suction blisters (elevated) or as punch biopsies. Each segment represents 5 samples. (C) Representative methylation profile of a human epidermis sample. Most markers were found to be unmethylated (beta<0.2), and a smaller group of markers was found to be highly methylated (beta>0.8). (D) DNA methylation profiles were compared between suction blister samples from two independent young donors. The results show a very high similarity with a correlation coefficient of r2 = 0.98. (E) Epidermal DNA methylation profiles were compared between two independent studies. Suction blister samples were obtained from male volunteers, punch biopsy samples were obtained from female individuals. Markers with major variations were almost completely localized to the X-chromosome (marked in blue), and can thus be attributed to the known hypermethylation of X-chromosomal loci in females.