Figure 5. Growth Data for MAL mutants.
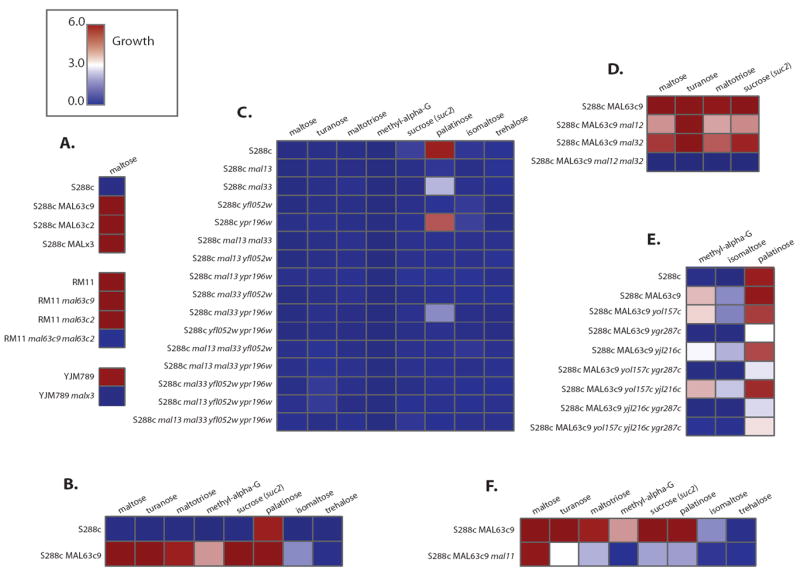
MAL deletion mutants confirm phenotypes and functional divergence. Growth of various MAL mutants is portrayed in a heatmap going from no growth (dark blue, 0.0) to strong growth (dark red, 6.0). A. S288c wild-type strain fails to grow on maltose while RM11 and YJM789 both grow on maltose. Transforming either a functional regulator from RM11 or YJM789 confers growth in S288c. Conversely, removing the functional regulator from YJM789, or both functional regulators from RM11 renders both strains unable to grow on maltose. B. The functional regulator from RM11, MAL63c9 (MAL63 found on supercontig 9 in RM11 (see Figure S5)), is not only required for growth on maltose, but is also required for growth on turanose, maltotriose, methy-α-glucoside, sucrose (suc2 mutant), palatinose, and isomaltose. C. All possible combinatorial knockouts of MAL regulators in S288c reveal that MAL13 and YFL052W are required for growth on palatinose, while the absence of MAL33 reduces growth on this carbon source. D. Two maltases, MALS genes, MAL12 and MAL32, are required for growth on maltose, turanose, maltotriose, and sucrose (the latter tested in a suc2 mutant), while the other MALS family members don’t affect the phenotype. E.YGR287C, a MALS gene, is the only MALS family member required for growth on palatinose, isomaltose, and methyl-α-glucoside. F. Removal of MAL11 permease renders strains unable to grow on most α-glucosides. See Figure S5 for more information.
