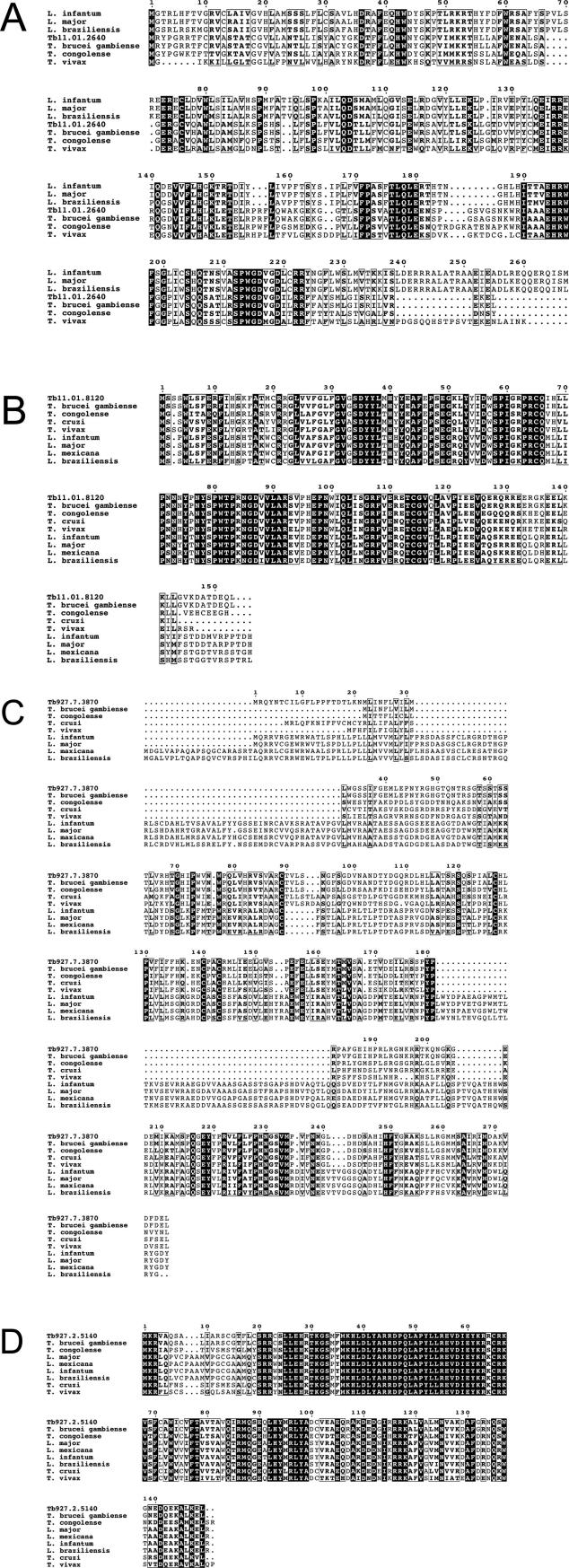
Supplementary Fig. 1.
Multiple sequence alignments of predicted amino acid sequences of four candidate ER proteins against their orthologues from selected Kinetoplastida genomes. Each homologue for each candidate was given the name based on the corresponding species name. Identical and similar amino acids are highlighted in black or outlined, respectively. A. Alignment for ERAP32 (Tb11.01.2640) orthologues across seven species. Accession numbers in vertical order: LinJ36_V3.5280, LmjF36.5050, LbrM35_V2.5300, Tb11.01.2640, gamb1662a11_1859d04, TcIL3000.11.11540, and Tviv1152c05.p1k_1. B. Alignment for ERAP18 (Tb11.01.8120) orthologues across nine species. Accession numbers in vertical order: Tb11.01.8120, Tbgamb.42801, TcIL3000.11.16350, Tc00.1047053506727.150, TvY486_1117370, LinJ32_V3.3890, LmjF32.3740, LmxM31.3740, LbrM32_V2.3980. C. Alignment of Tb927.7.3870 orthologues across nine species. Accession numbers in the vertical order: Tb927.7.3870, Tbgamb.14951, cIL3000.7.3130,Tc00.1047053511249.80, TvY486_0703860, LinJ14_V3.1150, LmjF14.1080, LmxM14.1080, LbrM14_V2.1070. D. Alignment of Tb927.2.5140 orthologues nine species. Accession numbers in vertical order: Tb927.2.5140, Tbgamb.2156, TcIL3000.2.1250, LmjF27.2370, LmxM27.2370, LinJ27_V3.2320, LbrM27_V2.2580, Tc00.1047053511627.80, tviv623d01.q1k_21.