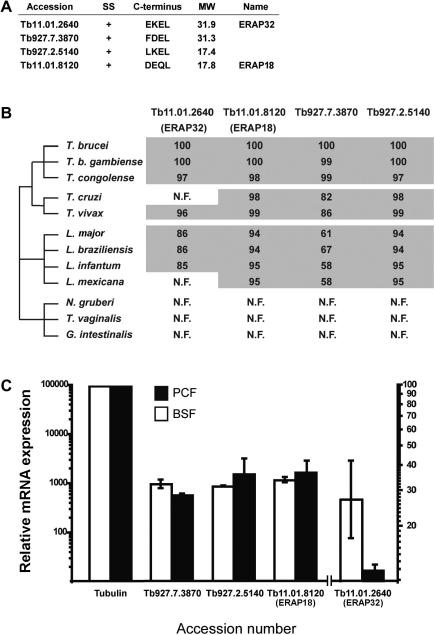
Fig. 1.
Identification and distribution of candidate novel ER proteins in Trypanosoma brucei. (A) GeneDB accession numbers and properties of the four candidate ER proteins selected for study. SS, signal peptide; C-terminus, C-terminal ER-extension like sequence; MW, molecular weight in kDa; +, N-terminal signal peptide predicted; Name, proposed name. (B) Distribution of candidate proteins across selected Excavata genomes. N.F. represents that evidence for an ortholog could not be found in the relevant taxon. Numbers represent the percent similarity by pairwise alignment with the T. brucei sequence as reference, using T-coffee. (C) mRNA expression levels of candidate ER proteins in PCF (black bars) and BSF (white bars) cells by qRT-PCR. Expression was normalized to tubulin. The experiment has been done a total of three times on different mRNA samples. Bars indicate the standard error.