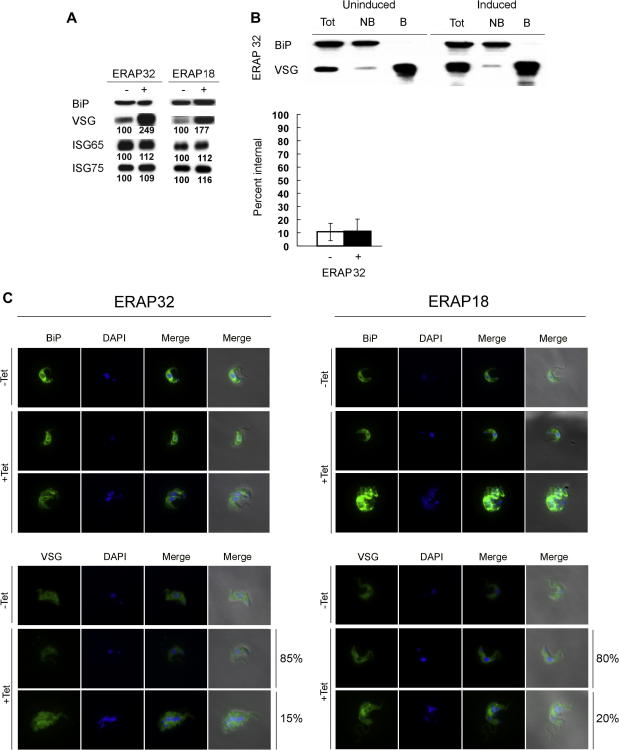
Fig. 4.
Alterations to VSG expression levels in ERAP32 and ERAP18 knockdown cells. (A) Western blot analysis of VSG abundance on RNAi induction. 1 × 107 cells were sampled from either induced (+) or uninduced (−) ERAP32 and ERAP18 RNAi cell lines after 1-day induction. Equivalent number of cells (1 × 106 cells) were applied to each lane. Primary antibody (rabbit anti-VSG221, rabbit anti-ISG75, rabbit anti-ISG65 or rabbit anti-BiP) was applied for 1 h at room temperature and secondary antibody was incubated with membrane for 40 min at room temperature. Pair numbers in each box indicate the relative abundance antigen in uninduced and induced cells from each RNAi cell line as determined by densitometry and normalized to the uninduced level. (B) Detection of VSG by surface biotinylation. Western blot is a representative biotinylation experiment performed on ERAP32 RNAi cell line at 12 h after induction. Tot: total VSG, NB: non-biotinylated, i.e. intracellular VSG, B: biotinylated, i.e. surface VSG. BiP was used as a loading and membrane integrity control. The lower graph represents quantification of VSG intensity from biotinylation assays following densitometry. Error bars represented mean ± standard error from at least two independent determinations. (C) IFA analysis of permeabilized cells silenced for ERAP32 and ERAP18. Upper panel: Visualization of ER structure and morphology of the cells in T. brucei stained with BiP. Lower panel: Distribution of VSG. Columns in each panel (from left to right): BiP/VSG stain (green); DAPI-stain of the nucleus and kinetoplast (blue); merged images with color combination for BiP/VSG and DAPI fluorescence; merged pictures from phase, DAPI and BiP/VSG. The first rows in each panel: uninduced cells (−Tet). The second and third rows in each panel are induced cells with normal morphology and abnormal morphology cells, respectively, and the percent representation of each morphology in the population indicated (cells with maintained normal morphology, and cells with defect in cell division and morphological abnormalities, respectively). Scale bar: 2 μm.