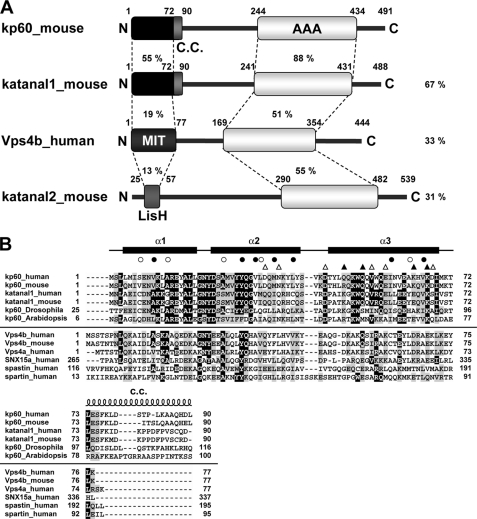
FIGURE 1.
Domain architectures and multiple sequence alignment of kp60s and proteins containing MIT domains. A, domain architectures of mouse kp60, katanal1 and -2, and human Vps4b. The amino acid identities of each domain and full-length proteins between kp60 and other proteins are indicated. C.C., coiled-coil; MIT, MIT domain; LisH, LIS1 homology domain; AAA, AAA domain. B, multiple sequence alignment of kp60-NTDs and related proteins with secondary structure elements of kp60-NTD. The secondary structure elements are shown at the top of the figure. The α-helices (α1–3) are represented as thick lines and the C.C. region as a coil. Filled and open circles above the alignments indicate well conserved and less conserved core residues, respectively (see Fig. 2A). Triangles indicate residues substituted with Ala for examining tubulin binding. (Filled triangle, involved in tubulin binding; open triangle, not involved.) Protein names and UniProtKB accession numbers are as follows: kp60 human (O75449); kp60 mouse (Q9WV86); kp60 Drosophila (Q9VN89); kp60 Arabidopsis (Q9SEX2); katanal1 human (Q9BW62); katanal1 mouse (Q8K0T4); Vps4b human (O75351); Vps4b mouse (P46467); Vps4a human (Q9UN37); SNX15a human (Q9NRS6); spartin human (Q8N0X7); and spastin human (Q9UBP0). The sequence alignment was generated by ClustalX (62).